✉Correspondences: 11email: zhiming.luo@xmu.edu.cn
Boundary Difference Over Union Loss For Medical Image Segmentation
Abstract
Medical image segmentation is crucial for clinical diagnosis. However, current losses for medical image segmentation mainly focus on overall segmentation results, with fewer losses proposed to guide boundary segmentation. Those that do exist often need to be used in combination with other losses and produce ineffective results. To address this issue, we have developed a simple and effective loss called the Boundary Difference over Union Loss (Boundary DoU Loss) to guide boundary region segmentation. It is obtained by calculating the ratio of the difference set of prediction and ground truth to the union of the difference set and the partial intersection set. Our loss only relies on region calculation, making it easy to implement and training stable without needing any additional losses. Additionally, we use the target size to adaptively adjust attention applied to the boundary regions. Experimental results using UNet, TransUNet, and Swin-UNet on two datasets (ACDC and Synapse) demonstrate the effectiveness of our proposed loss function. Code is available at https://github.com/sunfan-bvb/BoundaryDoULoss.
Keywords:
Medical image segmentation Boundary loss.1 Introduction
Medical image segmentation is a vital branch of image segmentation [16, 4, 23, 9, 15, 5], and can be used clinically for segmenting human organs, tissues, and lesions. Deep learning-based methods have made great progress in medical image segmentation tasks and achieved good performance, including early CNN-based methods [18, 25, 11, 10], as well as more recent approaches utilizing Transformers [24, 22, 8, 21, 7].
From CNN to Transformer, many different model architectures have been proposed, as well as a number of training loss functions. These losses can be mainly divided into three categories. The first class is represented by the Cross-Entropy Loss, which calculates the difference between the predicted probability distribution and the ground truth. Focal Loss [14] is proposed for addressing hard-to-learn samples. The second category is Dice Loss and other improvements. Dice Loss [17] is based on the intersection and union between the prediction and ground truth. Tversky Loss [19] improves the Dice Loss by balancing precision and recall. The Generalized Dice Loss [20] extends the Dice Loss to multi-category segmentation. The third category focuses on boundary segmentation. Hausdorff Distance Loss [12] is proposed to optimize the Hausdorff distance, and the Boundary Loss [13] calculates the distance between each point in the prediction and the corresponding ground truth point on the contour as the weight to sum the predicted probability of each point. However, the current loss for optimizing segmented boundaries dependent on combining different losses or training instability. To address these issues, we propose a simple boundary loss inspired by the Boundary IoU metrics [6], i.e., Boundary DoU Loss.
Our proposed Boundary DoU Loss improves the focus on regions close to the boundary through a region-like calculation similar to Dice Loss. The error region near the boundary is obtained by calculating the difference set of ground truth and prediction, which is then reduced by decreasing its ratio to the union of the difference set and a partial intersection set. To evaluate the performance of our proposed Boundary DoU loss, we conduct experiments on the ACDC [1] and Synapse datasets by using the UNet [18], TransUNet [3] and Swin-UNet [2] models. Experimental results show the superior performance of our loss when compared with others.
2 Method
This section first revisit the Boundary IoU metric [6]. Then, we describe the details of our Boundary DoU loss function and adaptive size strategy. Next, we discuss the connection between our Boundary DoU loss with the Dice loss.
2.1 Boundary IoU Metric
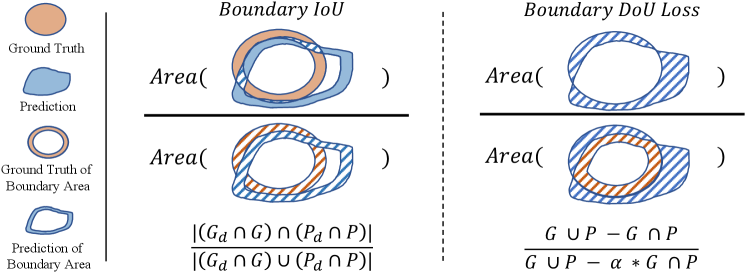
The Boundary IoU is a segmentation evaluation metric which mainly focused on boundary quality. Given the ground truth binary mask , the denotes the inner boundary region within the pixel width of . The is the predicted binary mask, and denotes the corresponding inner boundary region, whose size is determined as a fixed fraction of 0.5% relative to the diagonal length of the image. Then, we can compute the Boundary IoU metric by using following equation, as shown in the left of Fig. 1,
| (1) |
A large Boundary IoU value indicates that the and are perfectly matched, which means and are with a similar shape and their boundary are well aligned. In practice, the and is computed by the erode operation [6]. However, the erode operation is non-differentiable, and we can not directly leverage the Boundary IoU as a loss function for training for increasing the consistency between two boundary areas.
2.2 Boundary DoU Loss
As shown in the left of Fig. 1, we can find that the union of the two boundaries actually are highly correlated to the difference set between and . The intersection is correlated to the inner boundary of the intersection of and . If the difference set decreases and the increases, the corresponding Boundary IoU will increase.
Based on the above analysis, we design a Boundary DoU loss based on the difference region to facilitate computation and backpropagation. First, we directly treat the difference set as the miss-matched boundary between and . Besides, we consider removing the middle part of the intersection area as the inner boundary, which is computed by for simplicity. Then, we joint compute the as the partial union. Finally, as shown in the right of Fig. 1, our Boundary DoU Loss can be computed by,
| (2) |
where is a hyper-parameter controlling the influence of the partial union area.
Adaptive adjusting based-on target size: On the other aspect, the proportion of the boundary area relative to the whole target varies for different sizes. When the target is large, the boundary area only accounts for a small proportion, and the internal regions can be easily segmented, so we are encouraged to focus more on the boundary area. In such a case, using a large is preferred. However, when the target is small, neither the interior nor the boundary areas are easily distinguishable, so we need to focus simultaneously on the interior and boundary, and a small is preferred. To achieve this goal, we future adaptively compute based on the proportion,
| (3) |
where denotes the boundary length of the target, and denotes its size.
2.3 Discussion
In this part, we compare Boundary DoU Loss with Dice Loss. Firstly, we can re-write our Boundary DoU Loss as:
| (4) |
where denotes the area of the difference set between ground truth and prediction, denotes the intersection area of them, and . Meanwhile, the Dice Loss can be expressed by the following:
| (5) |
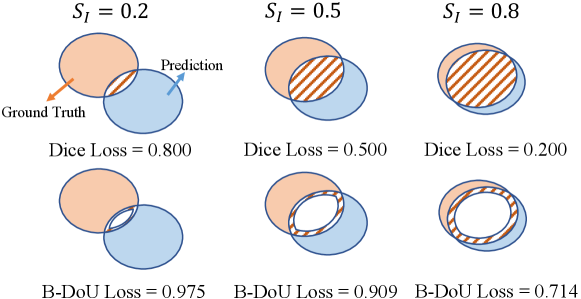
where TP, FP and FN denote True Positive, False Positive, and False Negative, respectively. It can be seen that Boundary DoU Loss and Dice loss differ only in the proportion of the intersection area. Dice is concerned with the whole intersection area, while Boundary DoU Loss is concerned with the boundary since . Similar to the Dice loss function, minimizing the will encourage an increase of the intersection area () and a decrease of the different set (). Meanwhile, the will penalize more over the ratio of . To corroborate its effectiveness more clearly, we compare the values of and in different cases in Fig. 2. The decreases linearly with the difference set, whereas will decrease faster when is higher enough.
3 Experiments
3.1 Datasets and Evaluation Metrics
Synapse:111https://www.synapse.org/#!Synapse:syn3193805/wiki/217789 The Synapse dataset contains 30 abdominal 3D CT scans from the MICCAI 2015 Multi-Atlas Abdomen Labeling Challenge. Each CT volume contains slices of pixels. The slice thicknesses range from 2.5 mm to 5.0 mm, and in-plane resolutions vary from to . Following the settings in TransUNet [3], we randomly select 18 scans for training and the remaining 12 cases for testing.
ACDC:222https://www.creatis.insa-lyon.fr/Challenge/acdc/ The ACDC dataset is a 3D MRI dataset from the Automated Cardiac Diagnosis Challenge 2017 and contains cardiac data from 150 patients in five categories. Cine MR images were acquired under breath-holding conditions, with slices 5-8 mm thick, covering the LV from basal to apical, with spatial resolutions from 1.37 to 1.68 mm/pixel and 28 to 40 images fully or partially covering the cardiac cycle. Following the TransUNet, we split the original training set with 100 scans into the training, validation, and testing sets with a ratio of 7:1:2.
Evaluation Metrics: We use the most widely used Dice Similarity Coefficient (DSC) and Hausdorff Distances (HD) as evaluation metrics. Besides, the Boundary IoU [6] (B-IoU) is adopted as another evaluation metric for the boundary.
| Model | UNet | TransUNet | Swin-UNet | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Loss | DSC | HD | B-IoU | DSC | HD | B-IoU | DSC | HD | B-IoU |
| Dice | 76.38 | 86.26 | 78.52 | 87.34 | 77.98 | 86.19 | |||
| CE | 65.95 | 82.69 | 72.98 | 84.84 | 71.77 | 84.04 | |||
| Dice + CE | 76.77 | 86.21 | 78.19 | 87.18 | 78.30 | 86.72 | |||
| Tversky | 63.61 | 75.65 | 63.90 | 70.77 | 68.22 | 79.53 | |||
| Boundary | 76.23 | 85.75 | 76.82 | 86.66 | 76.00 | 84.98 | |||
| Ours | 78.68 | 87.08 | 79.53 | 88.11 | 79.87 | 87.78 | |||
| Model | UNet | TransUNet | Swin-UNet | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Loss | DSC | HD | B-IoU | DSC | HD | B-IoU | DSC | HD | B-IoU |
| Dice | 90.17 | 75.20 | 90.69 | 76.66 | 90.17 | 75.20 | |||
| CE | 88.08 | 71.25 | 89.22 | 73.78 | 88.08 | 71.25 | |||
| Dice + CE | 89.94 | 74.80 | 90.48 | 76.27 | 89.94 | 74.80 | |||
| Tversky | 83.60 | 69.36 | 90.37 | 76.20 | 89.55 | 74.37 | |||
| Boundary | 89.25 | 73.08 | 90.48 | 76.31 | 88.95 | 72.73 | |||
| Ours | 90.84 | 76.44 | 91.29 | 78.45 | 91.02 | 77.00 | |||
| ACDC | Synapse | |||||||||||
| Model | UNet | TransUNet | Swin-UNet | UNet | TransUNet | Swin-UNet | ||||||
| Loss | large | small | large | small | large | small | large | small | large | small | large | small |
| Dice | 92.60 | 84.11 | 93.63 | 85.95 | 92.40 | 86.43 | 78.59 | 36.22 | 80.84 | 36.16 | 79.97 | 41.67 |
| CE | 91.40 | 81.72 | 92.55 | 83.86 | 91.49 | 82.58 | 68.89 | 0.12 | 75.21 | 32.32 | 74.27 | 26.25 |
| Dice + CE | 92.91 | 84.36 | 93.45 | 85.70 | 92.83 | 85.30 | 78.89 | 38.27 | 80.47 | 36.82 | 80.29 | 42.22 |
| Tversky | 91.82 | 70.37 | 93.28 | 85.86 | 92.67 | 84.53 | 65.76 | 24.60 | 66.00 | 25.74 | 70.08 | 34.44 |
| Boundary | 92.53 | 83.97 | 93.53 | 85.57 | 92.25 | 83.63 | 78.37 | 37.30 | 79.00 | 37.14 | 78.02 | 39.23 |
| Ours | 93.73 | 86.04 | 94.01 | 86.93 | 93.63 | 86.83 | 80.88 | 38.68 | 81.85 | 37.32 | 81.83 | 44.08 |
3.2 Implementation Details
We conduct experiments on three advanced models to evaluate the performance of our proposed Boundary DoU Loss, i.e., UNet, TransUNet, and Swin-UNet. The models are implemented with the PyTorch toolbox and run on an NVIDIA GTX A4000 GPU. The input resolution is set as for both datasets. For the Swin-UNet [2] and TransUNet [3], we used the same training and testing parameters provided by the source code, i.e., the learning rate is set to 0.01, with a weight decay of 0.0001. The batch size is 24, and the optimizer uses SGD with a momentum of 0.9. For the UNet, we choose ResNet50 as the backbone and initialize the encoder with the ImageNet pre-trained weights following the setting in TransUNet [3]. The other configurations are the same as TransUNet. We train all models by 150 epochs on both Synapse and ACDC datasets.
We further train the three models by different loss functions for comparison, including Dice Loss, Cross-Entropy Loss (CE), Dice+CE, Tversky Loss [19], and Boundary Loss [13]. The training settings of different loss functions are as follows. For the , we set as (0.5, 0.5) for the UNet and TransUNet, and (0.6, 0.4) for Swin-UNet. For the Tversky Loss, we set and by referring to the best performance in [19]. Following the Boundary Loss [13], we use for training . The is initially set to 1 and decreases by 0.01 at each epoch until it equals 0.01.

3.3 Results
Quantitative Results: Table 1 shows the results of different losses on the Synapse dataset. From the table, we can have the following findings: 1) The original Dice Loss achieves the overall best performance among other losses. The CE loss function obtains a significantly lower performance than the Dice. Besides, the Dice+CE, Tversky, and Boundary do not perform better than Dice. 2) Compared with the Dice Loss, our Loss improves the DSC by 2.30%, 1.20%, and 1.89% on UNet, TransUNet, and Swin-UNet models, respectively. The Hausdorff Distance also shows a significant decrease. Meanwhile, we achieved the best performance on the Boundary IoU, which verified that our loss could improve the segmentation performance of the boundary regions.
Table 2 reports the results on the ACDC dataset. We can find that our Boundary DOU Loss effectively improves DSC on all three models. Compared with Dice Loss, the DSC of UNet, TransUNet, and Swin-UNet improved by 0.62%, 0.6%, and 0.85%, respectively. Although our Loss did not get all optimal performance for the Hausdorff Distance, we substantially outperformed all other losses on the Boundary IoU. These results indicate that our method can better segment the boundary regions. This capability can assist doctors in better identifying challenging object boundaries in clinical settings.
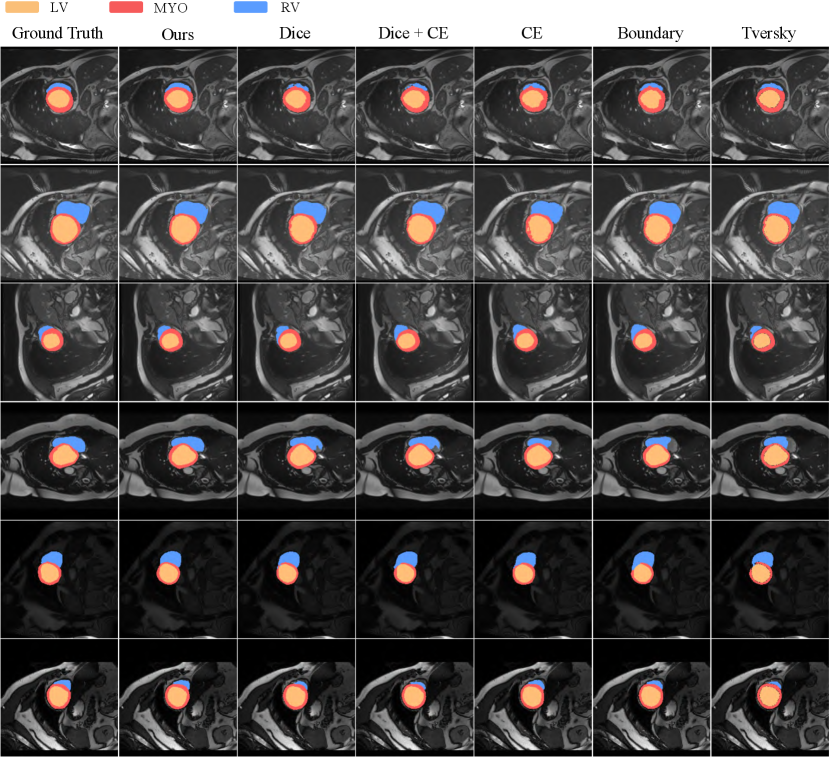
Qualitative Results: Fig. 3 and 4 show the qualitative visualization results of our loss and other losses. Overall, our method has a clear advantage for segmenting the boundary regions. In the Synapse dataset (Fig. 3), we can achieve more accurate localization and segmentation for complicated organs such as the stomach and pancreas. Our results from the 3rd and 5th rows substantially outperform the other losses when the target is small. Based on the 2nd and last rows, we can obtain more stable segmentation on the hard-to-segment objects. As for the ACDC dataset (Fig. 4), due to the large variation in the shape of the RV region as shown in Row 1, 3, 4 and 6, it is easy to cause under- or mis-segmentation. Our Loss resolves this problem better compared with other Losses. Whereas the MYO is annular and the finer regions are difficult to segment, as shown in the 2nd and 5th row, the other losses all result in different degrees of under-segmentation, while our loss ensures its completeness. Reducing the mis- and under-classification will allow for better clinical guidance.
Results of Target with Different Sizes: We further evaluate the influence of the proposed loss function for segmenting targets with different sizes. Based on the observation of values for different targets, we consider a target to be a large one when and otherwise as a small target. As shown in Table 3, our Boundary DoU Loss function can improve the performance for both large and small targets.
4 Conclusion
In this study, we propose a simple and effective loss (Boundary DoU) for medical image segmentation. It adaptively adjusts the penalty to regions close to the boundary based on the size of the different targets, thus allowing for better optimization of the targets. Experimental results on ACDC and Synapse datasets validate the effectiveness of our proposed loss function.
Acknowledgement
This work is supported by the National Natural Science Foundation of China (No. 62276221), the Natural Science Foundation of Fujian Province of China (No. 2022J01002), and the Science and Technology Plan Project of Xiamen (No. 3502Z20221025).
References
- [1] Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang, X., Heng, P.A., Cetin, I., Lekadir, K., Camara, O., Ballester, M.A.G., et al.: Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Transactions on Medical Imaging 37(11), 2514–2525 (2018)
- [2] Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., Wang, M.: Swin-unet: Unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537 (2021)
- [3] Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y.: Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
- [4] Chen, L.C., Papandreou, G., Schroff, F., Adam, H.: Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587 (2017)
- [5] Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: European Conference on Computer Vision (ECCV). pp. 801–818 (2018)
- [6] Cheng, B., Girshick, R., Dollár, P., Berg, A.C., Kirillov, A.: Boundary iou: Improving object-centric image segmentation evaluation. In: IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 15334–15342 (2021)
- [7] Gao, Y., Zhou, M., Metaxas, D.N.: Utnet: a hybrid transformer architecture for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 61–71. Springer (2021)
- [8] Hatamizadeh, A., Tang, Y., Nath, V., Yang, D., Myronenko, A., Landman, B., Roth, H.R., Xu, D.: Unetr: Transformers for 3d medical image segmentation. In: IEEE/CVF Winter Conference on Applications of Computer Vision. pp. 574–584 (2022)
- [9] He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask r-cnn. In: IEEE International Conference on Computer Vision. pp. 2961–2969 (2017)
- [10] Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q., Iwamoto, Y., Han, X., Chen, Y.W., Wu, J.: Unet 3+: A full-scale connected unet for medical image segmentation. In: IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). pp. 1055–1059. IEEE (2020)
- [11] Isensee, F., Petersen, J., Klein, A., Zimmerer, D., Jaeger, P.F., Kohl, S., Wasserthal, J., Koehler, G., Norajitra, T., Wirkert, S., et al.: nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv preprint arXiv:1809.10486 (2018)
- [12] Karimi, D., Salcudean, S.E.: Reducing the hausdorff distance in medical image segmentation with convolutional neural networks. IEEE Transactions on Medical Imaging 39(2), 499–513 (2019)
- [13] Kervadec, H., Bouchtiba, J., Desrosiers, C., Granger, E., Dolz, J., Ayed, I.B.: Boundary loss for highly unbalanced segmentation. In: International Conference on Medical Imaging with Deep Learning. pp. 285–296 (2019)
- [14] Lin, T.Y., Goyal, P., Girshick, R., He, K., Dollár, P.: Focal loss for dense object detection. In: IEEE International Conference on Computer Vision. pp. 2980–2988 (2017)
- [15] Liu, S., Qi, L., Qin, H., Shi, J., Jia, J.: Path aggregation network for instance segmentation. In: IEEE conference on Computer Vision and Pattern Recognition. pp. 8759–8768 (2018)
- [16] Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: IEEE Conference on Computer Vision and Pattern Recognition. pp. 3431–3440 (2015)
- [17] Milletari, F., Navab, N., Ahmadi, S.A.: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: International Conference on 3D Vision (3DV). pp. 565–571 (2016)
- [18] Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 234–241 (2015)
- [19] Salehi, S.S.M., Erdogmus, D., Gholipour, A.: Tversky loss function for image segmentation using 3d fully convolutional deep networks. In: International Workshop on Machine Learning in Medical Imaging. pp. 379–387 (2017)
- [20] Sudre, C.H., Li, W., Vercauteren, T., Ourselin, S., Jorge Cardoso, M.: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, pp. 240–248. Springer (2017)
- [21] Valanarasu, J.M.J., Oza, P., Hacihaliloglu, I., Patel, V.M.: Medical transformer: Gated axial-attention for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 36–46. Springer (2021)
- [22] Xu, G., Wu, X., Zhang, X., He, X.: Levit-unet: Make faster encoders with transformer for medical image segmentation. arXiv preprint arXiv:2107.08623 (2021)
- [23] Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: IEEE conference on computer vision and pattern recognition. pp. 2881–2890 (2017)
- [24] Zhou, H.Y., Guo, J., Zhang, Y., Yu, L., Wang, L., Yu, Y.: nnformer: Interleaved transformer for volumetric segmentation. arXiv preprint arXiv:2109.03201 (2021)
- [25] Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: Unet++: A nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, pp. 3–11. Springer (2018)