Brain Stroke Lesion Segmentation Using Consistent Perception Generative Adversarial Network
Abstract
The state-of-the-art deep learning methods have demonstrated impressive performance in segmentation tasks. However, the success of these methods depends on a large amount of manually labeled masks, which are expensive and time-consuming to be collected. In this work, a novel Consistent PerceptionGenerative Adversarial Network (CPGAN) is proposed for semi-supervised stroke lesion segmentation. The proposed CPGAN can reduce the reliance on fully labeled samples. Specifically, A similarity connection module (SCM) is designed to capture the information of multi-scale features. The proposed SCM can selectively aggregate the features at each position by a weighted sum. Moreover, a consistent perception strategy is introduced into the proposed model to enhance the effect of brain stroke lesion prediction for the unlabeled data. Furthermore, an assistant network is constructed to encourage the discriminator to learn meaningful feature representations which are often forgotten during training stage. The assistant network and the discriminator are employed to jointly decide whether the segmentation results are real or fake. The CPGAN was evaluated on the Anatomical Tracings of Lesions After Stroke (ATLAS). The experimental results demonstrate that the proposed network achieves superior segmentation performance. In semi-supervised segmentation task, the proposed CPGAN using only two-fifths of labeled samples outperforms some approaches using full labeled samples.
Index Terms:
Generative model, Semi-supervised learning, Consistent perception strategy, Stroke Lesion Segmentation.I Introduction
Stroke is a leading cause of dementia and depression worldwide[1]. Over two-thirds of stroke survivors experience long-term disabilities that impair their participation in daily activities[2, 3]. Stroke lesion segmentation is the first and essential step of lesion recognition and decision. Accurate identification and segmentation would improve the ability of physicians to correctly diagnose patients. Currently, the lesions are generally segmented manually by professional radiologists on MR images slice-by-slice, which is time-consuming and relies heavily on subjective perceptions[4]. Therefore, automatic methods for brain stroke lesion segmentation are in urgent demand in the clinical practice. Nevertheless, there are great challenges with this task. On the one hand, the scale, shape, size, and location of lesions limit the accuracy of automatic segmentation. On the other hand, some lesions have fuzzy boundaries, confusing the confidential partition between stroke and non-stroke regions.
With the development of machine learning in medical image analysis[5, 6, 7, 8, 9, 10], automatic feature learning algorithms have emerged as feasible approaches for stroke lesion segmentation. Zhang et al.[11] proposed a 3D fully convolutional and densely connected convolutional network (3D FC-DenseNet) for the accurate automatic segmentation of acute ischemic stroke. Bjoern at el.[12] designed a new generative probabilistic model for channel-specific tumor segmentation in multi-dimensional images. Hao et al.[13] designed a cross-level fusion and context inference network(CLCI-Net) for the chronic stroke lesion segmentation from T1-weighted MR images. Qi et al.[14] presented an end-to-end model named X-net for brain stroke lesion segmentation, this approach achieved good performance on ATLAS.
Although many automatic segmentation methods have been presented, they are essentially supervised learning methods. Training a robust model requires a large number of manually labeled masks. Due to the high cost for data labeling and patient privacy, it is difficult to collect sufficient samples for training of the model in medical image analysis. How to train an effective model using limited labeled data becomes an open and interesting problem.
The recent success of Generative Adversarial Networks(GANs)[15, 16, 17] and application of variational inference[18, 19, 20, 21] facilitates effective unsupervised learning in numerous tasks. The main reason is that GAN can automatically learn image characteristics in an unsupervised manner. Zhu et al.[22] designed an end-to-end adversarial FCN-CRF network for mammographic mass segmentation. Zhao et al. [23] proposed a cascaded generative adversarial network with deep-supervision discriminator (Deep-supGAN) for automatic bony structures segmentation. Lei et al. [24] adopted a effective GAN model for skin lesion segmentation from dermoscopy images. GAN has also been applied in the semi-supervised learning. For instance, Zhang et al.[25] proposed a novel semi-supervised method to check the coverage of LV from CMR images by using generative adversarial networks. Madani et al.[26] utilized a semi-supervised architecture of GANs to address both problems of labeled data scarcity and data domain overfitting. These studies have shown significant results by using both labeled data and arbitrary amounts of unlabeled data. However, the previous works focus mainly on the design of the generator and the use of fake samples, but fail to take full advantage of the discriminator and the data itself.
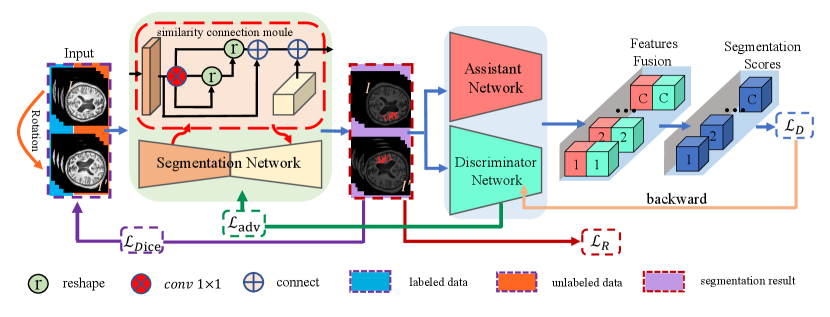
Motivated by this, we propose a novel method named Consistent Perception GAN(CPGAN) for semi-supervised segmentation task. A similarity connection module is designed in the segmentation network to capture the long-range contextual information, which contributes to the segmentation of lesions with different shapes and scales. This module can aggregate the information of multi-scale features and capture the spatial interdependencies of features. The assistant network is proposed to improve the performance of discriminator using meaningful feature representations. More importantly, A consistent perception strategy is developed in adversarial training. The rotation loss is adopted to encourage the segmentation network to make a consistent prediction of the input, which contains rotated and original images. A semi-supervised loss is designed according to the classification results of the discriminator and the assistant network. This loss can minimize the segmentation results between the labeled and unlabeled images.
In summary, the main contributions of this work can be listed as follows:
-
1.
A non-local operation, SCM is designed to capture context information from multi-scale features. The proposed SCM can selectively aggregate the features at each position and enhance the discriminant ability of the lesion areas.
-
2.
The assistant network is employed to encourage the discriminator to learn meaningful feature representations. The assistant network and discriminator work together and this structure can improve the performance of segmentation.
-
3.
A consistent perception strategy is proposed to improve the recognition of the unlabeled data. It makes full use of self-supervised information of the input and encourages the segmentation network to predict consistent results. Our method using only two-fifths of labeled samples outperforms some approaches using full labeled samples.
The rest of the paper is organized as follows. Section II discussed the relevant work about the architecture of the proposed network. Section III discussed the architecture and distinctive characteristics of CPGAN. Section IV summarized the results of extensive experiments including an ablation study for the similarity connection module and the assistant network. The quantitative and qualitative evaluations show that the proposed CPGAN achieves better performance of stroke lesion segmentation and has good performance of semi-supervised segmentation. Finally, Section V summarized the paper, and discusses some future research directions.
II Related Work
II-A U-net based methods
Encoder-decoder architectures based segmentation methods have been widely used in image segmentation task, such as U-net [27], H-DenseU-net [28],U-net++ [29]. It has becomes a popular neural network architecture for biomedical image segmentation tasks [30, 31, 32, 33]. Huang et al. [34] introduced a dense convolutional network (DenseNet) with dense blocks, which created short paths from the early layers to the latter layers. Baur et al. [35] proposed a semi-supervised learning framework for domain adaptation with embedding technique on challenging task of Multiple Sclerosis lesion segmentation. Sedai et al. [36] introduced a generative variational autoencoder that was trained using a limited number of labeled samples had a good performance of optic cup segmentation. Huang et al. [37] proposed a full-scale connected model Unet3 +, which has better segmentation performance for organs of different sizes in various medical images. However, existing models rely on an encoder-decoder architecture with stacked local operators to aggregate long-range information gradually. Those methods are easy to cause the loss of spatial information[38].
To address this issues, Ozan et al. [39] adopted a novel attention gate (AG) model for medical imaging that automatically learns to focus on target structures of varying shapes and sizes. Nabila et al. [40] combined attention gated U-Net with a novel variant of the focal Tversky loss function to address the problem of data imbalance in medical image segmentation. Wang et al. [41] introduced the non-local U-Nets equipped with flexible global aggregation blocks, this method outperforms previous models significantly with fewer parameters and faster computation on the 3D multimodality isointense infant brain MR image segmentation task. Fu et al. [42] proposed a dual attention network to integrate local features with global dependencies, their appended the position attention module and channel attention module on the top of FCN and achieved good performance on three challenging scene segmentation dataset. Unfortunately, those skip connections demand the fusion of the same-scale encoder and decoder feature maps[43], and those methods is insensitive to the different sizes and locations of lesions. Inspired by these previous studies, we proposed a segmentation module using a similarity connection module to enhance the ability of the representation in our CPGAN.
II-B GAN based methods
Generative Adversarial Networks(GANs), which can be derived via variational inference consist of two components parts: a generative network that generates pseudo data, and a discriminative network that differentiates between fake and real data. Recently, GAN has gained a lot of attention in the medical image computation field [7, 44, 45, 46, 47, 48] due to its capability of data generation. These properties have been adopted in many segmentation methods [49, 50, 51, 52, 53, 54, 55]. For example, Nie et al. [56] proposed a spatially-varying stochastic residual adversarial network (STRAINet) to delineate pelvic organs from MRI in an end-to-end fashion, this model achieved significant improvement in pelvic organ segmentation. Chen et al. [57] constructed a one-shot generative adversarial learning framework to make full use of both paired and unpaired MRI-CT data for MRI segmentation of craniomaxillofacial(CMF) bony structures. Micheal et al. [58] proposed a fully unsupervised segmentation approach exploiting image-to-image translation to convert from the image to the label domain. Xue et al. [59] proposed a novel end-to-end GAN, called SegAN, with a new multiscale loss for the task of brain tumor segmentation.
This adversarial training scheme and framework are also used for semi-supervised segmentation task. Chen et al.[60] proposed a semi-supervised method called MASSL that combines a segmentation task and a reconstruction task through an attention mechanism in a multi-task learning network. Zheng et al. [61] used adversarial learning with deep atlas prior to do semi-supervised segmentation of the liver in CT images. This method utilized unannotated data effectively and achieved good performance on ISBI LiTS 2017. However, the created pseudo labels usually do not have the same quality as the ground truth for the target segmentation objective, it would limit the performance of the semi-supervised segmentation models. Based on GAN structure, we introduce an assistant network and consistent perception strategy to improve the semi-supervised performance.
III Consistent Perception Generative Adversarial Network
The proposed semi-supervised learning method is shown in Fig. 1. CPGAN consists of three neural networks: segmentation network, discriminator network and assistant network. We adopt U-net architecture and equip skip connections with similarity connection module in the segmentation network to improve segmenting performance. Rotated images and original images input to the segmentation network to predict segmentation results. The network equivariant property[62] is utilized to obtain rotation loss and semi-supervised loss from labeled and unlabeled images.
III-A The architecture of Segmentation Network
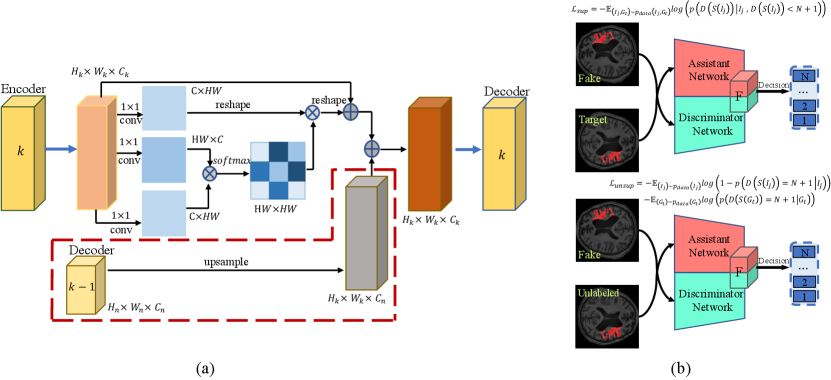
In the generator, U-net is applied to extract features. This architecture is composed of a down-sampling encoder and an up-sampling decoder. Skip connections are adopted to aggregate the same-scale feature map and capture local and global contextual information. However, a common limitation of the U-Net and its variations is that the consecutive pooling operations or convolution striding reduce the feature resolution to learn increasingly abstract feature representations. To address this challenge, a similarity connection module is proposed to extract a wide range of sensitive position information and multi-scale features. We append this non-local operation on the skip connections and sum the local information of varying scales at a decoder node.
The architecture of similarity connection module is shown in Fig.2(a). A local feature map is fed into a convolution layer which generates two new feature maps . Those feature maps are reshaped to , where and , is the number of layers in the Encoder. To compute the relationship of , for each pair of position, a matrix multiplication is calculated between and , consists of a softmax layer and measures the position’s impact on position:
| (1) |
Meanwhile, is fed into a convolution layer to generator a new feature map and reshaped to . Then, a matrix multiplication is calculated between and . The result is reshaped to . An upsample operation is performed on to get a new feature map , where . Finally, we sum and to obtain the output of similarity connection module:
| (2) |
Therefore, the result of similarity connection module can capture long-range contextual information according to non-local attention map and aggregate multi-scale feature to decrease the loss of spatial information.
III-B The architecture of Assistant Network
The original value function for GAN training is:
| (3) | ||||
Where is the true data distribution, is the generator’s distribution, and is the discriminator’s distribution. Training is typically performed via alternating stochastic gradient descent. Therefore, at iteration k during training, the discriminator classifies samples as coming from or . As the parameters of G change, the distribution changes, which implies a non-stationary online learning problem for the discriminator[63, 64]. To address this challenge, we propose an assistant network to prevent this forgetting of the classes in the discriminator representations.
As shown in Fig.2(b), the assistant network is pre-trained and shares the same architecture as the discriminator. Assistant network-parameters are fixed during the training, the predicted segmentation results input the assistant and discriminator network. We concatenate two feature maps, one from the last layer of the assistant network and the other from the discriminator. We can derive the gradient toward the discriminator by calculating the partial derivative of loss term :
| (4) | ||||
Where and is softmax function. and are network parameters. and represent assistant and discriminative feature maps, respectively. is gradient update formula of discriminator’s penultimate layer. Thus, depends on . The representative features affect the discriminator update. Therefore, the generator is trained by considering both assistant and discriminative features, because it should fool the discriminator by maximizing . Representative information of the assistant network would help the model to learn the information of stroke lesion faster and converge quickly.
III-C Training Strategy and Loss Functions
To improve the generalization capability of the network, the transformation equivariance has been proposed. Cohen and Welling [65] proposed group equivariant neural network to improve the network generalization. Dieleman et al. [66] designed four different equivariance to preserve feature map transformations by rotating feature maps instead of filters. Chen et al. [64] presented an unsupervised generative model that combines adversarial training with self-supervised learning by using auxiliary rotation loss. Inspired by these works, our consistent perception strategy targets to utilize the unlabeled images better in semi-supervised learning.
In the consistent perception strategy of the proposed method, labeled and unlabeled images are rotated. Rotated images and original images are added up to the input. Segmentation is desired as transformation equivariant. If the input images are rotated, the prediction of ground truth masks should be rotated in the same way compared to original masks. Rotation loss is adopted to evaluate the equivariant representation of segmentation network output on both labeled and unlabeled image, which is obtained by:
| (5) |
The segmentation network equipped with similarity connection module is trained using Dice loss on labeled images only:
| (6) |
Where and are cross-entropy loss and mean square error. is the ground truth label and is input images, and denote the segmentation network and rotation operation, and denotes the predicted results.
For unlabeled images, the rotation loss encourages the segmentation network to learn more self-supervised information. The adversarial loss brought by the discriminator provides a clever way of unlabeled samples into training. Some semi-supervised learning methods of GANs use discriminator for classification and treat generated data as . Instead, we exploit a novel technique to discriminate prediction of segmentation network. Ground truth, rotated and original labeled data are judged as , , , respectively. In this paper, .
Therefore, our loss function for training the discriminator is:
| (7) |
| (8) | ||||
| (9) | ||||
Meanwhile, we use the adversarial learning to improve the performance of segmentation. With the loss segmentation network is training to fool the discriminator by maximizing the probability of the prediction masks. This loss is generated from the distributions of ground truth masks:
| (10) |
In summary, the proposed rotation perception strategy training strategy encourages the discriminator to learn useful image representation and detects the rotated transformation. This semi-supervised training method promotes segmentation network to make the same prediction on both labeled and unlabeled images.
IV Experiments and Results
IV-A Data and Evaluation Metrics
IV-A1 Dataset
The CPGAN is evaluated on an open dataset, Anatomical Tracings of Lesions After Stroke (ATLAS), which contains 239 T1-weighted normalized 3D MR images with brain stroke lesion manually labeled mask. We randomly selected 139 subjects for training, 40 for validation and 60 for testing. Each of objects is cropped to 189 slices which size is ”233197” . In order to keep the size of rotated images consistent, slices are expended to ”256256” . Noted that only in the training dataset, the input images are rotated by .
IV-A2 Evaluation Metrics
In this paper, we employ 4-fold cross-validation strategy and use a series of evaluation metrics to measure the performance of our model, including Dice coefficient(Dic), Jaccard index(Jac), Accuracy(Acc), Sensitivity(Sen) and Specificity(Spe). The definition of them are:
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
Where ,, and refer to the number of true negatives, true positives, false negatives, false positives, respectively.
IV-A3 Implementation
Our implementation is based on Pytorch. 4 NVIDIA RTX 2080Ti with 11 GB memory are used for each experiment with a batch size of 4. The proposed network was trained with a fixed learning rate of 0.001. The strategy of reduce learning rate is adopted to reduce learning rate automatically and the Adam optimizer is used to minimize the loss function.
IV-B Ablation Analysis of Similarity Connection Module
We employ the similarity connection module(SCM) to extract a wide range of sensitive position information and multi-scale features. Three methods are used to conduct a comparative experiment. The similarity connection module is added into the architecture of U-net, ResUnet, and CPGAN, respectively. In Table I, it can be observed that employing SCM gains better performance in five evaluation metrics compared to the original method. Equipped with SCM, Unet-SCM(U-net with similarity connection module) performs better with 0.076, 0.061, 0.089, 0.053 and 0.116 improvement on Dice, Jaccard index, Accuracy, Sensitivity and Specificity, respectively. ResUnet-SCM(ResUnet with the similarity connection module) performs better with 0.069, 0.126, 0.060, 0.088 and 0.074 improvement on Dice, Jaccard index, Accuracy, Sensitivity and Specificity, respectively. Compared with Unet-SCM and ResUnet-SCM, similarity connection module is more effective in CPGAN with 0.103, 0.160, 0.109, 0.131 and 0.149 improvement on Dice, Jaccard index, Accuracy, Sensitivity and Specificity, respectively. All results show that the proposed module performs very well in Sensitivity and Specificity. It is worthwhile getting a higher score in these two evaluation metrics for brain stroke segmentation tasks, because we need to make sure that all the strokes can be detected and prevent non-diseased areas are misdiagnosed as brain stroke.
| Method | SCM | Dic | Jac | Acc | Sen | Spe |
|---|---|---|---|---|---|---|
| U-net | 0.468 | 0.374 | 0.542 | 0.440 | 0.573 | |
| ✓ | 0.544 | 0.435 | 0.631 | 0.493 | 0.689 | |
| ResUnet | 0.470 | 0.351 | 0.556 | 0.427 | 0.602 | |
| ✓ | 0.539 | 0.477 | 0.616 | 0.515 | 0.676 | |
| CPGAN | 0.514 | 0.421 | 0.529 | 0.425 | 0.556 | |
| ✓ | 0.617 | 0.581 | 0.638 | 0.556 | 0.705 |
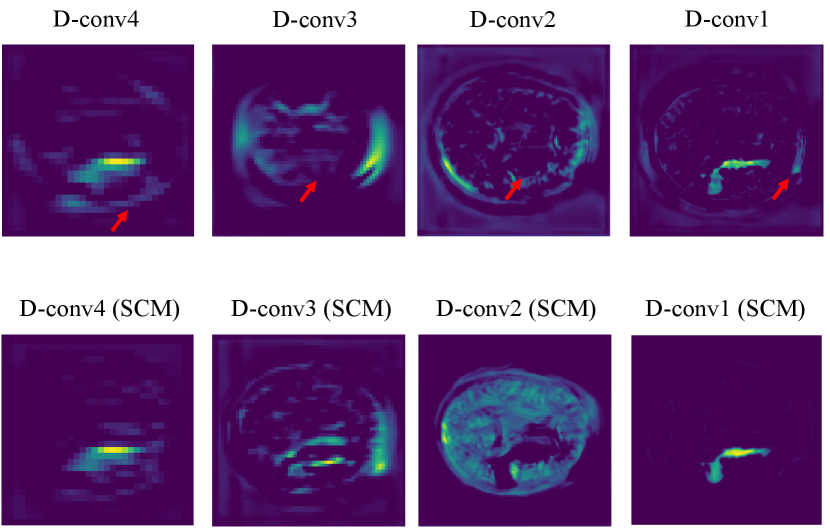
To understand the advantages of the similarity connection module better, we further present the visualization results of the features from the Decoder part of CPGAN unequipped with SCM in Fig.3. From this figure, the original Decoder part has two problems. For one thing, model learns redundant information in non-lesion areas. For another, the valid information is not captured in lesion areas. Compared with the CPGAN. SCM has ability of capturing long-range contextual information of the stroke lesion areas and reducing the learning of the redundant information.
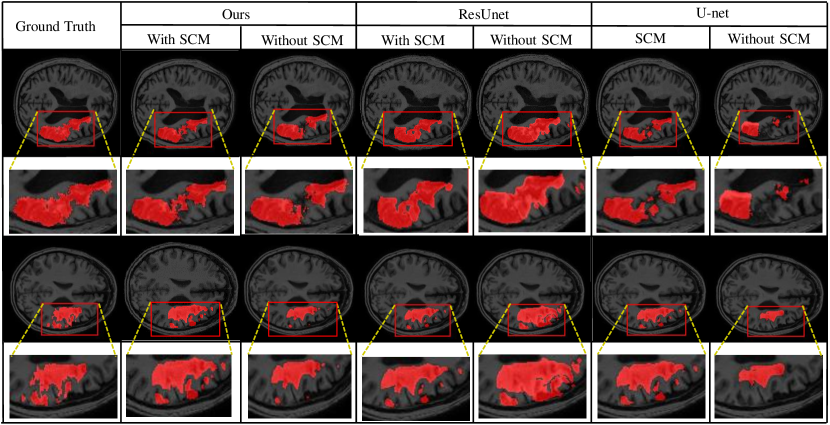
The segmentation results of different methods are shown in Fig.4. More details are captured with our proposed similarity connection module. It is demonstrated that the proposed module can help model achieve better performance of segmentation consistently, and some of the interdependencies might have already been captured with our proposed SCM.
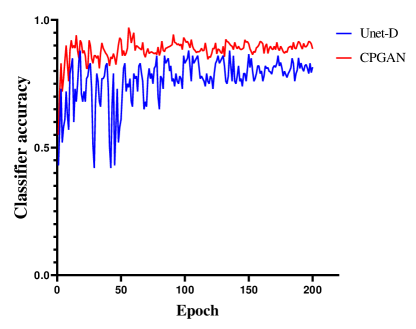
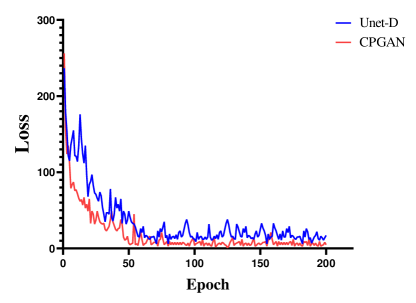
IV-C Analysis of Assistant Network
A set of experiments are conducted to validate the effectiveness of the proposed assistant network. To verify the effectiveness of this network, we conduct experiments with two models: CPGAN, Unet-D unequipped with the assistant network, the rest of the Unet-D’s structure is the same as CPGAN. It could be clearly observed from Fig.5 and Fig.6 that employing assistant network enhances the performance of the model. Fig.5 shows the change of the classification accuracy of the discriminator. The Unet-D is unstable from 0 to 50 epochs. The classification accuracy drops substantially every 5 epochs. After 150 epochs, the classification accuracy is beginning to stabilize. This demonstrates that the discriminator does not retain useful information in this non-stationary environment. Some representations of lesion areas are forgotten during training and this forgetting correlates with training instability. After adding assistant network, we observe that proposed method can mitigate this problem. Compared with Unet-D, the classification accuracy of CPGAN is stable after 20 epochs and the performance improves by an average of 10 percent. Representative information of assistant network improves the discriminator to learn meaningful feature representations. It can be verified that assistant network improves performance of CPGAN. As shown in Fig.6, we can observe that the loss of segmentation network changes. CPGAN decreases faster than Unet-D. After 50 epochs, the loss of CPGAN tends to be stable, while Unet-D requires 70 epochs. This experiment indicates that adopting the proposed assistant network can help discriminator mitigate the problem of discriminator forgetting. It can be inferred that our proposed method converges quickly and achieves better performance.
IV-D Rotation Perception strategy Strategy and Semi-supervised segmentation
| Method | Dic | Jac | Acc | Sen | Spe |
|---|---|---|---|---|---|
| DeepLab V3+[67] | 0.487 | 0.392 | 0.571 | 0.523 | 0.585 |
| Dense U-net[27] | 0.538 | 0.466 | 0.628 | 0.562 | 0.657 |
| U-net[27] | 0.468 | 0.374 | 0.542 | 0.440 | 0.573 |
| DCGAN[68] | 0.439 | 0.388 | 0.529 | 0.425 | 0.556 |
| X-Net[14] | 0.572 | 0.457 | 0.646 | 0.493 | 0.679 |
| CPGAN | 0.617 | 0.581 | 0.638 | 0.556 | 0.705 |
IV-D1 Comparison to state-of-the-art methods
We compare CPGAN with different state-of-the-art segmentation methods, including U-net, DenseU-net(2D), DeepLab V3+[67], DCGAN[68] and X-net[14]. We briefly introduce these models here and the details can be found in the references. DCGAN applies the convolutional operators to replace the pooling operators, strided convolutions for the discriminator and fractional strided convolutions for the generator. X-net adds a feature similarity module and a X-block to the U-net based architecture, it is the top three methods on ATALS dataset leaderboard. From the results listed in Table I, it can be clearly observed that the proposed model scores are the highest on the main indicators. Besides, Table II shows that our CPGAN performs better than other methods and our segmentation method makes significant improvement than [14]. Although our method is not as good as other methods in Accuarcy and sensitivity, it delivers promising performance on other evaluation metrics. Compared with X-net, our method performs better with 0.045, 0.124 and 0.026 improvement on Dice, Jaccard and specificity, respectively.
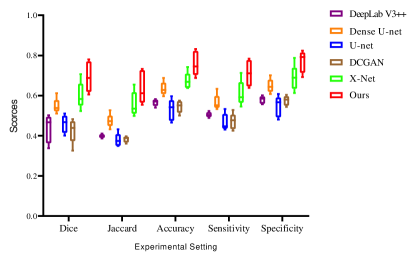
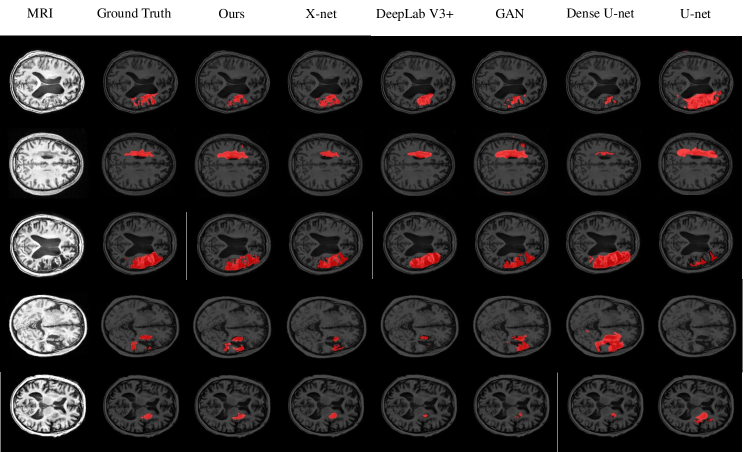
To further evaluate our model, we divide the testing set into 5 parts to draw the box-plots. Fig.7 shows the box-plots of Dice, Jaccard index, accuracy, sensitivity, and specificity of different models. These results demonstrate that the performance of our model is superior to other methods. Some details of the segmentation results are shown in Fig.8. It can be inferred that our proposed CPGAN can segment the brain stroke lesions in T1-weighted MR images very well. Our model performs well on some fuzzy lesion boundaries and the confidential partition between stroke and non-stroke regions. It is very important to help specialists measure the stroke in stroke segmentation tasks.
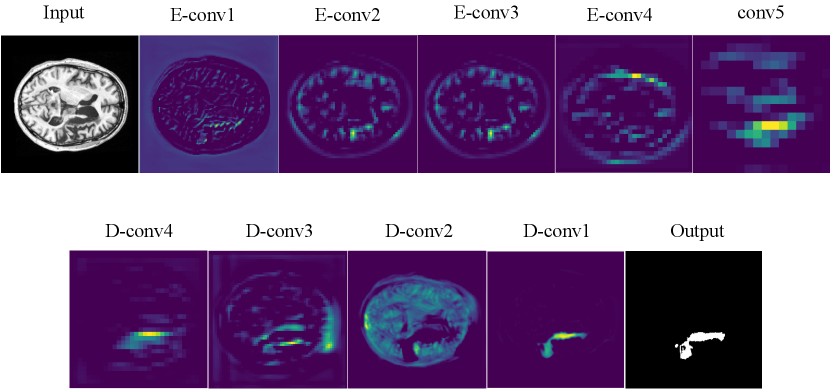
To better understand what features our model has learned, each of layers in the CPGAN are visualized in Fig.10. From these feature visualizations, our proposed CPGAN can capture the important pixel areas of stroke lesion in T1-weight MR images. It demonstrates that our model has excellent ability in stroke lesion segmentation.
IV-D2 Effectiveness of semi-supervised segmentation
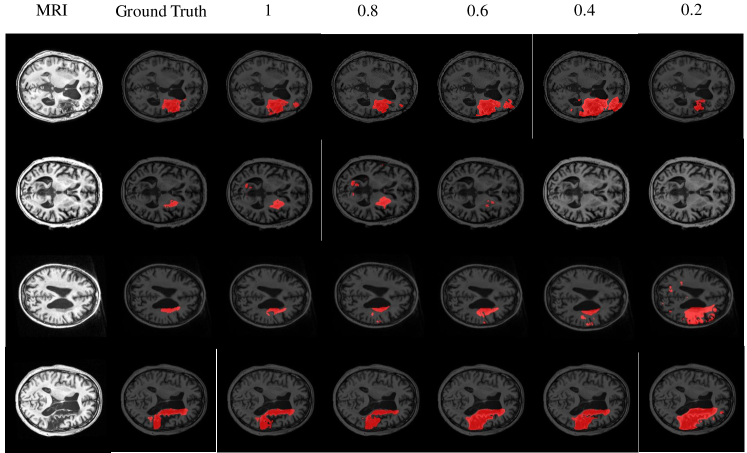
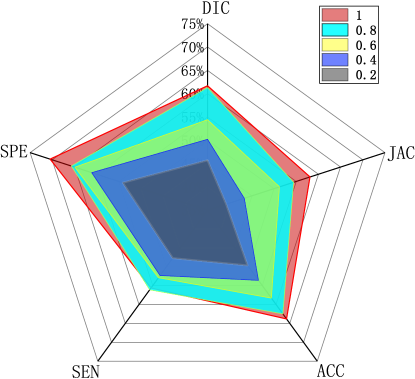
To show the semi-supervised segmentation performance of CPGAN, 5 experiments are designed and the number of labeled images are different for each set of experiments. The rate of labeled images is set to 1, 0.8, 0.6, 0.4 and 0.2. As shown in the TableIII and Fig.11, our semi-supervised method performs better than some full supervised method in different labeled/unlabeled data settings, which demonstrates that our method effectively utilizes unlabeled data and proposed method is beneficial to the performance gains. Only three-fifths of labeled images are used, our proposed model performs better than X-net in Table with 0.055 improvement on Jaccard index. Furthermore, when the rate is set to two-fifths, the scores of Dice, Jaccard, sensitivity and specificity are higher than U-net. The comparison shows the effectiveness of our semi-supervised segmentation method achieves the training effect of some full supervised methods. Some semi-supervised segmentation results of different rates are shown in Fig.9. It can be inferred that the proposed method presented stronger capability in semi-supervised brain stroke lesion segmentation.
| Labeled/Full | Dic | Jac | Acc | Sen | Spe |
|---|---|---|---|---|---|
| 1 | 0.617 | 0.581 | 0.638 | 0.556 | 0.705 |
| 0.8 | 0.613 | 0.544 | 0.625 | 0.531 | 0.657 |
| 0.6 | 0.544 | 0.512 | 0.583 | 0.529 | 0.649 |
| 0.4 | 0.502 | 0.433 | 0.536 | 0.523 | 0.611 |
| 0.2 | 0.457 | 0.392 | 0.496 | 0.477 | 0.541 |
V Conclusion
In this paper, we propose a novel semi-supervised segmentation method named CPGAN for brain stroke lesion segmentation. The similarity connection module is adopted into the segmentation network. The effectiveness of the proposed similarity connection module is verified through ablation study. This module can effectively improve the details of the lesion area of segmentation by capturing long-range spatial information. The proposed assistant network is pre-trained and shares the same architecture as discriminator. The hyper-parameters of assistant network are fixed during the training. The qualitative and quantitative experimental results demonstrate that representative information of this network help discriminator mitigate the problem of discriminator forgetting and improve performance of segmentation network. The proposed consistent Perception strategy strategy is very useful to semi-supervised segmentation. Only two-fifths of labeled images are used, our proposed model performs better than some other methods. Results suggest that our method performs better in segmentation on the ATLAS dataset. This method can also be extended to other medical image segmentation tasks. In this work, we only experimented with one change of angle and found this transformation strategy is very useful for semi-supervised segmentation. In the future, we will explore the application of the consistent perception strategy and add more transformations to improve the performance of semi-supervised segmentation.
Acknowledgments
This work was supported by the National Natural Science Foundations of China under Grants 62172403 and 61872351, the International Science and Technology Cooperation Projects of Guangdong under Grant2019A050510030, the Distinguished Young Scholars Fund of Guangdong under Grant 2021B1515020019, the Excellent Young Scholars of Shenzhen under Grant RCYX20200714114641211 and Shenzhen KeyBasic Research Project under Grants JCYJ20180507182506416 and JCYJ20200109115641762.
References
- [1] W. Johnson, O. Onuma, M. Owolabi, and S. Sachdev, “Stroke: a global response is needed.” Bulletin of the World Health Organization, vol. 94, no. 9, p. 634, 2016.
- [2] V. L. Feigin, M. H. Forouzanfar, R. Krishnamurthi, G. A. Mensah, M. Connor, D. A. Bennett, A. E. Moran, R. L. Sacco, L. Anderson, T. Truelsen et al., “Global and regional burden of stroke during 1990–2010: findings from the global burden of disease study 2010,” The Lancet, vol. 383, no. 9913, pp. 245–255, 2014.
- [3] G. Kwakkel, B. J. Kollen, J. van der Grond, and A. J. Prevo, “Probability of regaining dexterity in the flaccid upper limb: impact of severity of paresis and time since onset in acute stroke,” Stroke, vol. 34, no. 9, pp. 2181–2186, 2003.
- [4] S.-L. Liew, J. M. Anglin, N. W. Banks, M. Sondag, K. L. Ito, H. Kim, J. Chan, J. Ito, C. Jung, N. Khoshab et al., “A large, open source dataset of stroke anatomical brain images and manual lesion segmentations,” Scientific data, vol. 5, p. 180011, 2018.
- [5] S. Wang, H. Wang, Y. Shen, and X. Wang, “Automatic recognition of mild cognitive impairment and alzheimers disease using ensemble based 3d densely connected convolutional networks,” in 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA). IEEE, 2018, pp. 517–523.
- [6] S. Wang, Y. Shen, C. Shi, P. Yin, Z. Wang, P. W.-H. Cheung, J. P. Y. Cheung, K. D.-K. Luk, and Y. Hu, “Skeletal maturity recognition using a fully automated system with convolutional neural networks,” IEEE Access, vol. 6, pp. 29 979–29 993, 2018.
- [7] S. Wang, Y. Hu, Y. Shen, and H. Li, “Classification of diffusion tensor metrics for the diagnosis of a myelopathic cord using machine learning,” International journal of neural systems, vol. 28, no. 02, p. 1750036, 2018.
- [8] S. Wang, Y. Shen, D. Zeng, and Y. Hu, “Bone age assessment using convolutional neural networks,” in 2018 International Conference on Artificial Intelligence and Big Data (ICAIBD). IEEE, 2018, pp. 175–178.
- [9] S. Wang, X. Wang, Y. Shen, B. He, X. Zhao, P. W.-H. Cheung, J. P. Y. Cheung, K. D.-K. Luk, and Y. Hu, “An ensemble-based densely-connected deep learning system for assessment of skeletal maturity,” IEEE Transactions on Systems, Man, and Cybernetics: Systems, 2020.
- [10] S. Yu, S. Wang, X. Xiao, J. Cao, G. Yue, D. Liu, T. Wang, Y. Xu, and B. Lei, “Multi-scale enhanced graph convolutional network for early mild cognitive impairment detection,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020, pp. 228–237.
- [11] R. Zhang, L. Zhao, W. Lou, J. M. Abrigo, V. C. Mok, W. C. Chu, D. Wang, and L. Shi, “Automatic segmentation of acute ischemic stroke from dwi using 3-d fully convolutional densenets,” IEEE transactions on medical imaging, vol. 37, no. 9, pp. 2149–2160, 2018.
- [12] B. H. Menze, K. Van Leemput, D. Lashkari, T. Riklin-Raviv, E. Geremia, E. Alberts, P. Gruber, S. Wegener, M.-A. Weber, G. Székely et al., “A generative probabilistic model and discriminative extensions for brain lesion segmentation—with application to tumor and stroke,” IEEE transactions on medical imaging, vol. 35, no. 4, pp. 933–946, 2015.
- [13] H. Yang, W. Huang, K. Qi, C. Li, X. Liu, M. Wang, H. Zheng, and S. Wang, “Clci-net: Cross-level fusion and context inference networks for lesion segmentation of chronic stroke,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 266–274.
- [14] K. Qi, H. Yang, C. Li, Z. Liu, M. Wang, Q. Liu, and S. Wang, “X-net: Brain stroke lesion segmentation based on depthwise separable convolution and long-range dependencies,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 247–255.
- [15] I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, and Y. Bengio, “Generative adversarial nets,” Advances in neural information processing systems, vol. 27, pp. 2672–2680, 2014.
- [16] S. Hu, B. Lei, S. Wang, Y. Wang, Z. Feng, and Y. Shen, “Bidirectional mapping generative adversarial networks for brain mr to pet synthesis,” IEEE Transactions on Medical Imaging, 2021.
- [17] W. Yu, B. Lei, Y. Liu, Z. Feng, Y. Hu, Y. Shen, S. Wang, and M. K. Ng, “Morphological feature visualization of alzheimer’s disease via multidirectional perception gan,” IEEE Transactions on Neural Networks and Learning Systems, 2021.
- [18] L.-F. Mo and S.-Q. Wang, “A variational approach to nonlinear two-point boundary value problems,” Nonlinear Analysis: Theory, Methods & Applications, vol. 71, no. 12, pp. e834–e838, 2009.
- [19] S.-Q. Wang, “A variational approach to nonlinear two-point boundary value problems,” Computers & Mathematics with Applications, vol. 58, no. 11-12, pp. 2452–2455, 2009.
- [20] S.-Q. Wang and J.-H. He, “Variational iteration method for a nonlinear reaction-diffusion process,” International Journal of Chemical Reactor Engineering, vol. 6, no. 1, 2008.
- [21] ——, “Variational iteration method for solving integro-differential equations,” Physics letters A, vol. 367, no. 3, pp. 188–191, 2007.
- [22] W. Zhu, X. Xiang, T. D. Tran, G. D. Hager, and X. Xie, “Adversarial deep structured nets for mass segmentation from mammograms,” in 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018). IEEE, 2018, pp. 847–850.
- [23] M. Zhao, L. Wang, J. Chen, D. Nie, Y. Cong, S. Ahmad, A. Ho, P. Yuan, S. H. Fung, H. H. Deng et al., “Craniomaxillofacial bony structures segmentation from mri with deep-supervision adversarial learning,” in International conference on medical image computing and computer-assisted intervention. Springer, 2018, pp. 720–727.
- [24] B. Lei, Z. Xia, F. Jiang, X. Jiang, Z. Ge, Y. Xu, J. Qin, S. Chen, T. Wang, and S. Wang, “Skin lesion segmentation via generative adversarial networks with dual discriminators,” Medical Image Analysis, vol. 64, p. 101716, 2020.
- [25] L. Zhang, A. Gooya, and A. F. Frangi, “Semi-supervised assessment of incomplete lv coverage in cardiac mri using generative adversarial nets,” in International Workshop on Simulation and Synthesis in Medical Imaging. Springer, 2017, pp. 61–68.
- [26] A. Madani, M. Moradi, A. Karargyris, and T. Syeda-Mahmood, “Semi-supervised learning with generative adversarial networks for chest x-ray classification with ability of data domain adaptation,” in 2018 IEEE 15th International symposium on biomedical imaging (ISBI 2018). IEEE, 2018, pp. 1038–1042.
- [27] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241.
- [28] X. Li, H. Chen, X. Qi, Q. Dou, C.-W. Fu, and P.-A. Heng, “H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes,” IEEE transactions on medical imaging, vol. 37, no. 12, pp. 2663–2674, 2018.
- [29] Z. Zhou, M. M. R. Siddiquee, N. Tajbakhsh, and J. Liang, “Unet++: A nested u-net architecture for medical image segmentation,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, 2018, pp. 3–11.
- [30] M. Dunnhofer, M. Antico, F. Sasazawa, Y. Takeda, S. Camps, N. Martinel, C. Micheloni, G. Carneiro, and D. Fontanarosa, “Siam-u-net: encoder-decoder siamese network for knee cartilage tracking in ultrasound images,” Medical Image Analysis, vol. 60, p. 101631, 2020.
- [31] Y. Hiasa, Y. Otake, M. Takao, T. Ogawa, N. Sugano, and Y. Sato, “Automated muscle segmentation from clinical ct using bayesian u-net for personalized musculoskeletal modeling,” IEEE Transactions on Medical Imaging, 2019.
- [32] Y. Man, Y. Huang, J. Feng, X. Li, and F. Wu, “Deep q learning driven ct pancreas segmentation with geometry-aware u-net,” IEEE transactions on medical imaging, vol. 38, no. 8, pp. 1971–1980, 2019.
- [33] Q. Zhu, B. Du, and P. Yan, “Boundary-weighted domain adaptive neural network for prostate mr image segmentation,” IEEE Transactions on Medical Imaging, 2019.
- [34] G. Huang, Z. Liu, L. Van Der Maaten, and K. Q. Weinberger, “Densely connected convolutional networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 4700–4708.
- [35] C. Baur, S. Albarqouni, and N. Navab, “Semi-supervised deep learning for fully convolutional networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp. 311–319.
- [36] S. Sedai, D. Mahapatra, S. Hewavitharanage, S. Maetschke, and R. Garnavi, “Semi-supervised segmentation of optic cup in retinal fundus images using variational autoencoder,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp. 75–82.
- [37] H. Huang, L. Lin, R. Tong, H. Hu, Q. Zhang, Y. Iwamoto, X. Han, Y.-W. Chen, and J. Wu, “Unet 3+: A full-scale connected unet for medical image segmentation,” in ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE, 2020, pp. 1055–1059.
- [38] Z. Gu, J. Cheng, H. Fu, K. Zhou, H. Hao, Y. Zhao, T. Zhang, S. Gao, and J. Liu, “Ce-net: context encoder network for 2d medical image segmentation,” IEEE transactions on medical imaging, vol. 38, no. 10, pp. 2281–2292, 2019.
- [39] O. Oktay, J. Schlemper, L. L. Folgoc, M. Lee, M. Heinrich, K. Misawa, K. Mori, S. McDonagh, N. Y. Hammerla, B. Kainz et al., “Attention u-net: Learning where to look for the pancreas,” arXiv preprint arXiv:1804.03999, 2018.
- [40] N. Abraham and N. M. Khan, “A novel focal tversky loss function with improved attention u-net for lesion segmentation,” in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE, 2019, pp. 683–687.
- [41] Z. Wang, N. Zou, D. Shen, and S. Ji, “Non-local u-nets for biomedical image segmentation,” in Thirty-Fourth AAAI Conference on Artificial Intelligence, 2020.
- [42] J. Fu, J. Liu, H. Tian, Y. Li, Y. Bao, Z. Fang, and H. Lu, “Dual attention network for scene segmentation,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2019, pp. 3146–3154.
- [43] S. Hasan and C. A. Linte, “U-netplus: a modified encoder-decoder u-net architecture for semantic and instance segmentation of surgical instrument,” arXiv preprint arXiv:1902.08994, 2019.
- [44] S. Wang, X. Wang, Y. Hu, Y. Shen, Z. Yang, M. Gan, and B. Lei, “Diabetic retinopathy diagnosis using multichannel generative adversarial network with semisupervision,” IEEE Transactions on Automation Science and Engineering, 2020.
- [45] W. Yu, B. Lei, M. K. Ng, A. C. Cheung, Y. Shen, and S. Wang, “Tensorizing gan with high-order pooling for alzheimer’s disease assessment,” IEEE Transactions on Neural Networks and Learning Systems, 2021.
- [46] S. Hu, Y. Shen, S. Wang, and B. Lei, “Brain mr to pet synthesis via bidirectional generative adversarial network,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020, pp. 698–707.
- [47] S. You, Y. Liu, B. Lei, and S. Wang, “Fine perceptive gans for brain mr image super-resolution in wavelet domain,” arXiv preprint arXiv:2011.04145, 2020.
- [48] S. Hu, W. Yu, Z. Chen, and S. Wang, “Medical image reconstruction using generative adversarial network for alzheimer disease assessment with class-imbalance problem,” in 2020 IEEE 6th International Conference on Computer and Communications (ICCC). IEEE, 2020, pp. 1323–1327.
- [49] J. M. Wolterink, T. Leiner, M. A. Viergever, and I. Išgum, “Generative adversarial networks for noise reduction in low-dose ct,” IEEE transactions on medical imaging, vol. 36, no. 12, pp. 2536–2545, 2017.
- [50] S. U. Dar, M. Yurt, L. Karacan, A. Erdem, E. Erdem, and T. Çukur, “Image synthesis in multi-contrast mri with conditional generative adversarial networks,” IEEE transactions on medical imaging, vol. 38, no. 10, pp. 2375–2388, 2019.
- [51] Q. Yang, P. Yan, Y. Zhang, H. Yu, Y. Shi, X. Mou, M. K. Kalra, Y. Zhang, L. Sun, and G. Wang, “Low-dose ct image denoising using a generative adversarial network with wasserstein distance and perceptual loss,” IEEE transactions on medical imaging, vol. 37, no. 6, pp. 1348–1357, 2018.
- [52] B. Yu, L. Zhou, L. Wang, Y. Shi, J. Fripp, and P. Bourgeat, “Ea-gans: edge-aware generative adversarial networks for cross-modality mr image synthesis,” IEEE transactions on medical imaging, vol. 38, no. 7, pp. 1750–1762, 2019.
- [53] A. Sharma and G. Hamarneh, “Missing mri pulse sequence synthesis using multi-modal generative adversarial network,” IEEE transactions on medical imaging, vol. 39, no. 4, pp. 1170–1183, 2019.
- [54] Michael, Gadermayr, Laxmi, Gupta, Vitus, Appel, Peter, Boor, Barbara, and M. and, “Generative adversarial networks for facilitating stain-independent supervised and unsupervised segmentation: A study on kidney histology.” IEEE Transactions on Medical Imaging, 2019.
- [55] M. T. Chen, F. Mahmood, J. A. Sweer, and N. J. Durr, “Ganpop: Generative adversarial network prediction of optical properties from single snapshot wide-field images,” IEEE Transactions on Medical Imaging, 2019.
- [56] D. Nie, L. Wang, Y. Gao, J. Lian, and D. Shen, “Strainet: Spatially varying stochastic residual adversarial networks for mri pelvic organ segmentation,” IEEE transactions on neural networks and learning systems, vol. 30, no. 5, pp. 1552–1564, 2018.
- [57] X. Chen, C. Lian, L. Wang, H. Deng, S. H. Fung, D. Nie, K.-H. Thung, P.-T. Yap, J. Gateno, J. J. Xia et al., “One-shot generative adversarial learning for mri segmentation of craniomaxillofacial bony structures,” IEEE transactions on medical imaging, 2019.
- [58] M. Gadermayr, L. Gupta, V. Appel, P. Boor, B. M. Klinkhammer, and D. Merhof, “Generative adversarial networks for facilitating stain-independent supervised and unsupervised segmentation: A study on kidney histology,” IEEE transactions on medical imaging, vol. 38, no. 10, pp. 2293–2302, 2019.
- [59] Y. Xue, T. Xu, H. Zhang, L. R. Long, and X. Huang, “Segan: Adversarial network with multi-scale l 1 loss for medical image segmentation,” Neuroinformatics, vol. 16, no. 3-4, pp. 383–392, 2018.
- [60] S. Chen, G. Bortsova, A. G.-U. Juárez, G. van Tulder, and M. de Bruijne, “Multi-task attention-based semi-supervised learning for medical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 457–465.
- [61] H. Zheng, L. Lin, H. Hu, Q. Zhang, Q. Chen, Y. Iwamoto, X. Han, Y.-W. Chen, R. Tong, and J. Wu, “Semi-supervised segmentation of liver using adversarial learning with deep atlas prior,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 148–156.
- [62] D. E. Worrall, S. J. Garbin, D. Turmukhambetov, and G. J. Brostow, “Harmonic networks: Deep translation and rotation equivariance,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 5028–5037.
- [63] D. Bang and H. Shim, “Improved training of generative adversarial networks using representative features,” arXiv preprint arXiv:1801.09195, 2018.
- [64] T. Chen, X. Zhai, M. Ritter, M. Lucic, and N. Houlsby, “Self-supervised gans via auxiliary rotation loss,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, pp. 12 154–12 163.
- [65] T. S. Cohen and M. Welling, “Group equivariant convolutional networks,” arXiv: Learning, 2016.
- [66] S. Dieleman, J. De Fauw, and K. Kavukcuoglu, “Exploiting cyclic symmetry in convolutional neural networks,” arXiv: Learning, 2016.
- [67] L.-C. Chen, Y. Zhu, G. Papandreou, F. Schroff, and H. Adam, “Encoder-decoder with atrous separable convolution for semantic image segmentation,” in Proceedings of the European conference on computer vision (ECCV), Cham, 2018, pp. 801–818.
- [68] Z. Li, Y. Wang, and J. Yu, “Brain tumor segmentation using an adversarial network,” in International MICCAI brainlesion workshop. Cham: Springer, 2017, pp. 123–132.