Computational Reproducibility Within Prognostics and Health Management
Abstract
Scientific research frequently involves the use of computational tools and methods. Providing thorough documentation, open-source code, and data – the creation of reproducible computational research – helps others understand a researcher’s work. Here, we explore computational reproducibility, broadly, and from within the field of prognostics and health management (PHM). The adoption of reproducible computational research practices remains low across scientific disciplines and within PHM. Our text mining of more than 300 articles, from publications engaged in PHM research, showed that fewer than 1% of researchers made both their code and data available to others. Although challenges remain, there are also clear opportunities, and personal benefits, for engaging in reproducible computational research. Highlighting an opportunity, we introduce an open-source software tool, called PyPHM, to assist PHM researchers in accessing and preprocessing common industrial datasets.
von Hahn
1 Introduction
An article about computational science […] is merely advertising of the scholarship. The actual scholarship is the complete software development environment and the complete set of instructions which generated the figures.
The above statement, provocatively expressed by Buckheit and Donoho [Buckheit \BBA Donoho (\APACyear1995)] paraphrases the thoughts of Jon Claebout. Claebout, a geophysicist, became an early advocate for reproducible computational research. Simply put, reproducible computational research is performed when “all details of the computation — code and data — are made conveniently available to others.” [Donoho \BOthers. (\APACyear2008)]
Claebout’s advocacy, in the early 1990s, came at a time when computation was ascending as a means of conducting scientific research. Researchers were wrestling with how these new tools affected the dissemination of ideas. Today, computational research is ubiquitous across a multitude of fields. In addition, the paradigms of the internet, immense computational power, massive data, and open-source software, have enabled tremendous scientific advances. Yet, creating and encouraging reproducible computational research remains a challenge [Ince \BOthers. (\APACyear2012)], [\APACciteatitleTrouble at the lab (\APACyear2013)].
Prognostics and health management (PHM) “is an enabling technology used to maintain the reliable, efficient, economic and safe operation of engineering equipment, systems and structures.” [Hu \BOthers. (\APACyear2022)] PHM practitioners build these technologies with the same computational tools used across the breadth of modern science. As such, the field encounters similar challenges surrounding reproducible computational research.
The work presented here explores the topic of reproducible computational research from within the context of PHM. We ask four questions:
-
1.
What does reproducible computational research look like? We present a practical example from PHM.
-
2.
What is the state of reproducible computational research? We look at findings from the broader scientific community and discuss our own findings from within PHM.
-
3.
What are the benefits to conducting reproducible computational research? We discuss the underappreciated personal benefits of conducting reproducible computational research. Namely, performing computationally reproducible research yields greater exposure of one’s work, stronger career opportunities, and increased personal satisfaction.
-
4.
What are the challenges and opportunities for reproducible computational research? We segment the challenges and opportunities into three categories – experience, motivation, and resources. We discuss them broadly and from within PHM.
In addition, we introduce an open-source software package, called PyPHM111PyPHM is publicly available on GitHub: https://github.com/tvhahn/PyPHM , to assist PHM practitioners in accessing and understanding public domain datasets and creating reproducible data workflows. Other fields have similar software packages, and we note that there is need of one within PHM. As such, we invite others to assist in this endeavor.
Isaac Newton wrote to his fellow scientist, Robert Hooke, that “if I have seen further, it is by standing on the shoulders of giants.” Newton then passed on his ideas through his published writings and texts. However, for future generations to stand on our shoulders we should pass on more than writings. The data and the code are also needed.
2 Example of Reproducible Computational Research
PHM methods can be categorized into physics based and data-driven methods. Work that uses the physics of failure, to produce a PHM application, may require the code to be available if other researchers are to reproduce it. However, in a data-driven approach, both the code and data are needed to reproduce the work. The data-driven approach will be the focus in this section.
Figure 1, as an example, shows the simplified steps used to create a machine learning model for detecting tool wear on a CNC machine. The figure, combined with Table 1, highlights some considerations for making the work computationally reproducible.
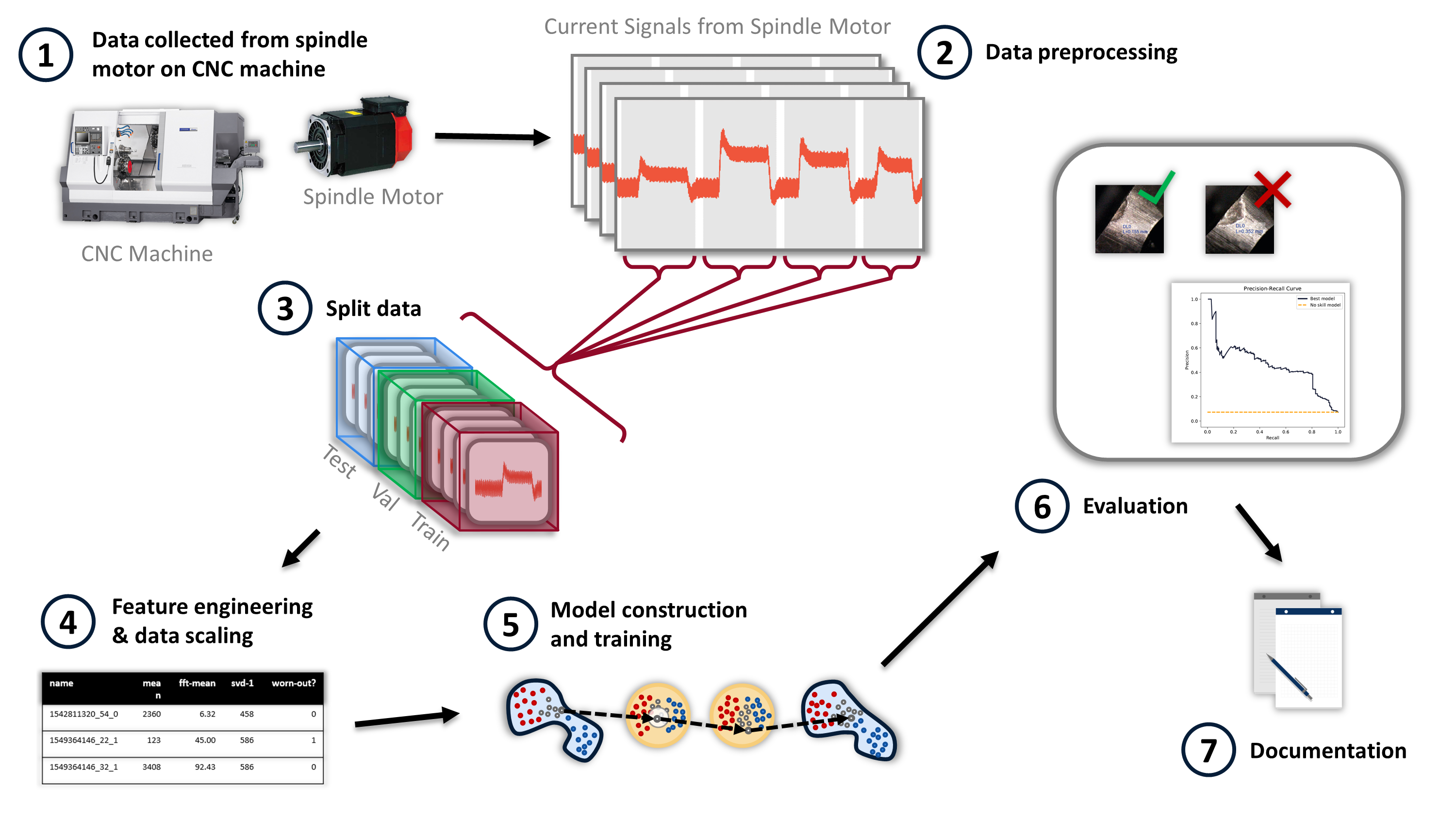
| Step | Description | Considerations for Computational Reproducibility | ||||
| 1 | Data Collection |
|
||||
| 2 | Data Preprocessing |
|
||||
| 3 | Data Splits |
|
||||
| 4 | Feature Engineering and Data Scaling |
|
||||
| 5 | Model Construction and Training |
|
||||
| 6 | Evaluation |
|
||||
| 7 | Documentation |
|
Table 1, below, illustrates the complexities of reproducible computational research. Modern data-driven research contains intricate data preprocessing steps, feature engineering, and a complicated selection of parameters. Rarely can the details of this computational workflow be fully conveyed in a research paper. Consequently, the code, data, and additional documentation are needed to access this tacit knowledge. Unfortunately, many researchers only document their work through published papers.
3 State of Reproducible Computational Research
The National Academies of Sciences, Engineering, and Medicine detailed the state of computational reproducibility in science in their 2019 report [National Academies of Sciences, Engineering, and Medicine and others (\APACyear2019)]. Their work showed that the lack of computational reproducibility is still a concern across science. For example, a study in computational physics demonstrated that only 6% of articles, from 307 surveyed, make the data and code available [Stodden \BOthers. (\APACyear2018)].
Within the AI research domain, Gunderson et al. [Gundersen \BOthers. (\APACyear2018)] found that fewer than 6% of articles (out of a sample of 400) provided access to their code, and fewer than 30% used a dataset that was publicly available. François Chollet, the creator of Keras, a popular deep learning library, laments that many deep learning papers today are “often optimized for peer review in both style and content in ways that actively hurt clarity of explanation and reliability of results.” [Chollet (\APACyear2021)] Unfortunately, this “optimization”, as expressed by Chollet, does not encourage computational reproducibility.
Does computational reproducibility fair better within PHM? Astfalck et al. surveyed 50 PHM papers between 2000 and 2014, focusing on papers building data-driven prognostic models [Astfalck \BOthers. (\APACyear2016)]. Only eight of the papers (16%) utilized open-source or readily available datasets, and only one paper (2%) had the code available for inspection.
We too sought an understanding of computational reproducibility within PHM. As such, we text mined approximately 375 articles from three venues. The venues – Energies, Mechanical Signals and Systems Processing (MSSP), and the PHM Conference – all feature work, to varying degrees, from within the PHM domain.
In the text mining process we searched for keywords and context that indicated whether the data or code, used in the research, was publicly available. As a comparison, we also text mined 100 computer science and electrical engineering articles from arXiv, an open-access archive of scholarly articles. All the articles, from across the four venues, were randomly sampled and drawn between the years of 2015 and 2021.
Figure 2 demonstrates the text mining process for a single publication venue. The process was implemented in Python using open-source libraries. The interested reader is encouraged to visit the project page222This work, and our future text mining research, is being conducted in the open and is available on GitHub: https://github.com/tvhahn/arxiv-code-search and inspect the code of this text mining process.
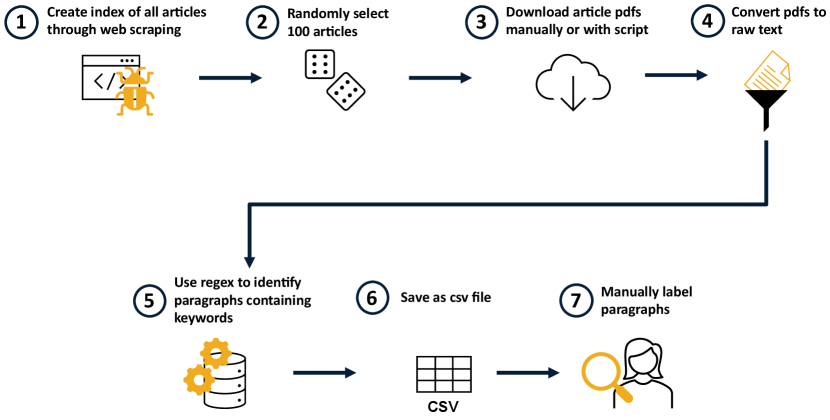
In the first step, as illustrated in Figure 2, web-scraping was used to build an index of all the papers published, at a specified venue, during a certain period. The web-scraping libraries Beautiful Soup and Selenium were used. From there, 100 to 150 articles were randomly selected. The pdfs of the articles were then downloaded with a script, or manually, depending on the publication venue.
In step four, the raw text from the pdfs were extracted using pdfminer.six. Regular expressions (regex) were implemented using Python’s standard library to search for keywords and short phrases, as shown in step five. Table 2 demonstrates several regular expressions, amongst many, that were used in the keyword search. If a keyword match was found then the paragraph containing the keyword was saved into a csv file.
| Keywords | Regex Code |
| “used dataset” | \b(used)(?:\W+\w+){0,5}?\W+(dataset)\b |
| “open-source” | \b(open-source|open source)\b |
| “code available” | \b(code)(?:\W+\w+){0,9}?\W+(available) |
Finally, each paragraph in the csv file was manually labelled to designate if the paragraph provided indication of public data or code. The results were then aggregated across each unique article.
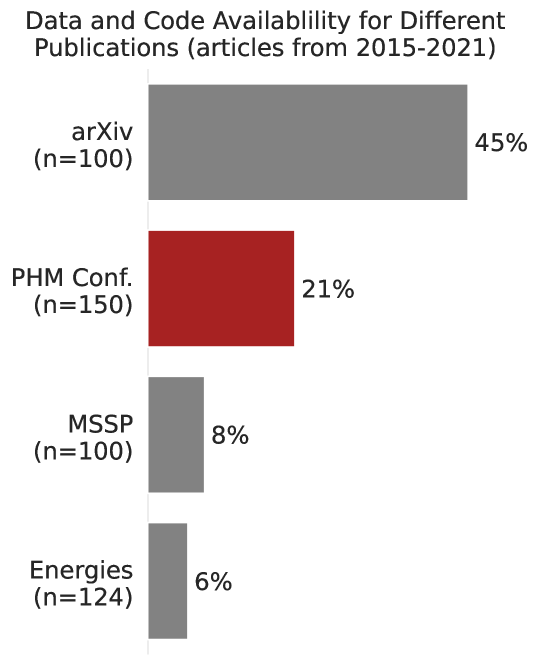
From the results of the text mining, as shown in Figure 3, 21% of the technical papers from the PHM Conference provided public access to their data or code. However, only 4% of the articles sampled from the PHM Conference, as shown in Figure 4, provide access to the code used in their research. In general, we observed that data is much more likely to be made publicly available than the code, regardless of the publication venue. The distributions from the other publication venues are found in Figure 7 in the Appendix.
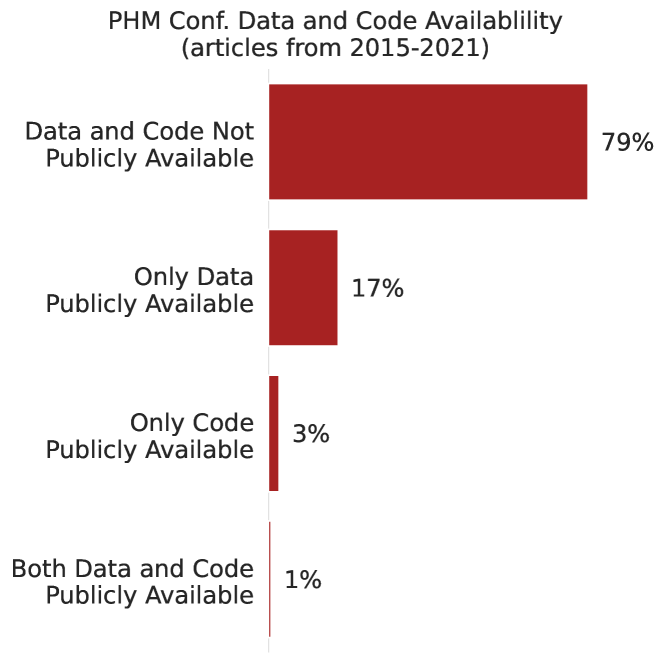
Our text mining effort is ongoing as we seek a broader understanding of computational reproducibility within PHM and beyond, and we will further document these results in future publications. For now, we believe this observational study corroborates the evidence from Astfalck et al.; that is, the field of PHM suffers from similar issues of computational reproducibility as in other disciplines.
4 Personal Benefits of Reproducible Computational Research
We observe that many commentators, when discussing computational reproducibility, appeal to the reader’s sense of altruism and morality. “Reproducibility is a cornerstone of the scientific method” [Gundersen \BOthers. (\APACyear2018)] and therefore, it should be honored. In fact, we too strongly appealed to the reader’s sense of “rightness” in the introduction.
However, in this section, we highlight several benefits of computationally reproducible work that are less discussed. Rather than appealing to a sense of altruism, these appeal to the individual’s self-interest. Namely, reproducible computational research can lead to increased exposure of a researcher’s work, better career opportunities, and a greater sense of satisfaction.
4.1 Increased Exposure
Reproducible computational research, by its nature, requires work to be open and transparent. Most often, this necessitates that the code, data, and text are made freely available on the internet. Fortunately, this additional effort does not go unrewarded.
A significant amount of research has shown that freely releasing the code, data, and published text (either through a preprint or open-access article) leads to increased citations \shortcitefrachtenberg2022research,dorch2015data,henneken2011linking,piwowar2013data,piwowar2007sharing,colavizza2020citation,fu2019meta,christensen2019study. In some domains, the increase was 2-fold \shortcitewahlquist2018dissemination. Including the code and data, alongside scientific articles, is a clear and obvious way to produce differentiated research.
4.2 Career Opportunities
STEM (science, technology, engineering, and mathematics) occupations are “projected to grow over two times faster than the total for all occupations in [this] decade,” according to the U.S. Bureau of Labor Statistics. Computer related occupations will produce most of this growth [Zilberman \BBA Ice (\APACyear2021)]. Researchers engaged in computational science will stand to benefit as their skills are increasingly in demand.
In this competitive job market, employers have begun to accept alternate credentials, as opposed to a traditional university degrees. Candidates can be hired through an intensive bootcamp program or enter a company as an apprentice. Competency can also be demonstrated through a real-world portfolio of work [Rainie \BBA Anderson (\APACyear2017)].
Creating computationally reproducible research requires the full body of work to be accessible and understandable. Therefore, not only does reproducible computational research help the scientific community, but it also creates a strong body of work for an individual’s portfolio, thus enhancing their opportunities for employment.
4.3 Satisfaction
The phenomenon of open-source software (OSS) consistently raises one question: why do individuals dedicate enormous amounts of their time for little economic benefit? Intrinsic motivation – a sense of internal satisfaction – is seen as a strong driver for individuals to contribute to OSS [Hars \BBA Ou (\APACyear\bibnodate)], [Bitzer \BOthers. (\APACyear2007)].
The act of producing reproducible research is like that of open-source software development. One’s work, through the code, data, and documents, is given to the world with no expectation of reward. Yet, the act of creating the work, and then selflessly sharing it with others, is internally satisfying. From the personal experience of the authors, this is one benefit of reproducible computational research that should not be ignored.
5 Challenges and Opportunities
As discussed above, there are challenges in creating reproducible computational research. We have split these challenges, and subsequent opportunities, into three categories, as shown in Figure 5.
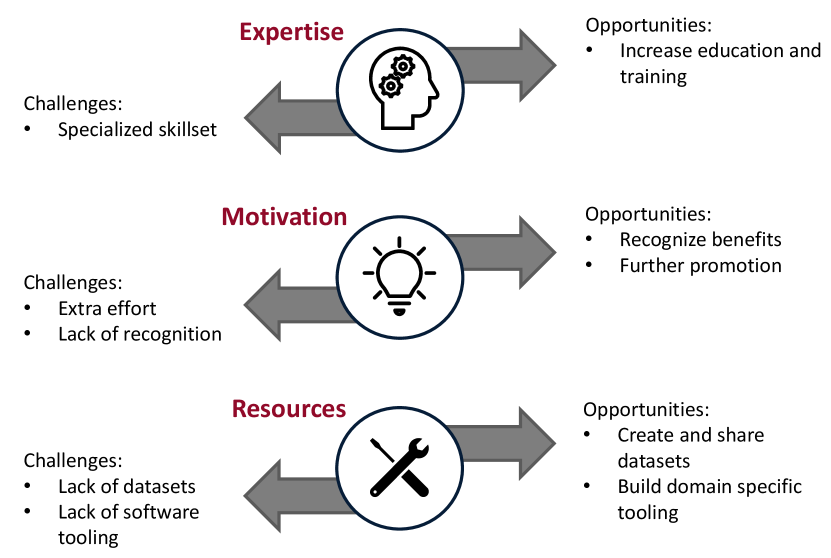
The first challenge concerns expertise. Creating reproducible computational research requires a specialized skillset that is generally covered in less depth by university curricula [National Academies of Sciences, Engineering, and Medicine and others (\APACyear2019)]. The skillset may include the use of version control, for both code and data; knowledge of containerization; or expertise in Linux, to name a few examples.
Education efforts are being made to improve computational researcher’s expertise. Software Carpentry, a volunteer run organization, has been offering training since 1998 to improve the computational skillset of researchers [Wilson (\APACyear2014)]. Topics of computational reproducibility have been added to the curriculum, from medicine to computer science, at multiple universities [\APACcitebtitleAn introduction to the foundations of neuro data science (\APACyear\bibnodate), \APACcitebtitleReproducible and Collaborative Data Science (\APACyear\bibnodate), \APACcitebtitlePrinciples, Statistical and Computational Tools for Reproducible Data Science (\APACyear2017)]. However, we are unaware of any courses or training specific to PHM. This is an area of opportunity.
The second challenge concerns that of motivation. Reproducible computational research requires extra effort. Unfortunately, the extra effort, combined with the pressure to publish and lack of recognition, creates an impediment to reproducibility [National Academies of Sciences, Engineering, and Medicine and others (\APACyear2019)]. As discussed in Section 4, we believe that a wider recognition towards the benefits of reproducible computational research can help motivate researchers. Funding organizations, and academic journals, are also encouraging researchers to consider computational reproducibility [Stodden \BOthers. (\APACyear2013)].
Likewise, PHM specific conferences and journals should include measures to encourage computational reproducibility. As an example of this encouragement, a measure of computational reproducibility can be integrated into the peer review process. Specific recognition can also be given to papers that demonstrate computational reproducibility. Overall, a broader discussion on how to improve computational reproducibility is warranted.
Finally, the lack of domain specific resources, either in tooling or publicly available datasets, can impede computational reproducibility. The computational workflow, within a specific field, may be similar across a variety of research projects. Thus, software tools can be created to standardize these workflows. The standardization allows researchers to better grasp and more quickly reproduce each other’s work and avoids a myriad of ad-hoc approaches. The standardization also enables researchers to spend more time on higher-value tasks, such as developing novel algorithms, as opposed to preprocessing data. The software can also be coupled with open-source datasets to facilitate the comparison of results between research groups.
Table 3, below, lists several open-source software packages that are specific to certain domains. These software packages assist researchers in accessing datasets and reproducing computational workflows. The software, and their documentation, also assists in educating researchers on domain specific problems, techniques, and methods, and demonstrate how to implement solutions in a reproducible manner.
| Software Name | Domain | Description |
| fMRIPrep \shortciteesteban2019fmriprep | Neuroimaging | Preprocessing pipeline for functional-MRI data |
| medicaltorch \shortcitemedicaltorchPerone | Medical imaging | General package for accessing medical imaging datasets and standardize preprocessing methods |
| astroML \shortciteastroML | Astronomy and astrophysics | Machine learning tools and data for astronomy and astrophysics |
| torchvision \shortciteNEURIPS2019_9015 | Computer vision | Popular datasets, model architectures, and image transformations for computer vision. |
| Natural Language Toolkit (NLTK) \shortciteBird_Natural_Language_Processing_2009 | Natural language processing | Open-source modules, datasets, and tutorials supporting research and development in natural language processing |
As an example of this software, consider torchvision. The software allows researchers to download common computer vision datasets, apply well recognized preprocessing techniques in a standardized way, and even load already trained deep learning models. Various tutorials and examples are available to help individuals understand the functionality of the software package.
Within PHM, there is a noted lack of high-quality, and large, datasets [Zhao \BOthers. (\APACyear2019)], [Wang \BOthers. (\APACyear2021)]. The lack of these datasets may be due to the proprietary nature of industrial data or the poor understanding of their need within the PHM research community. We encourage others to freely share their PHM datasets.
The field of PHM, to the knowledge of the authors, also lacks an open-source software package, like those found in Table 3. Such a tool would enable the easy access to PHM datasets and the implementation of computational workflows. We see this as an opportunity, and as such, we have begun the process of building such a software tool, discussed below.
6 An Open-Source Software Tool for PHM Datasets
Currently, PHM datasets are spread-out across the internet and require users to both find the data and then manually download it. Furthermore, users must implement their own data preprocessing steps, which can be a time-consuming endeavor. The open-source software tool being developed, called PyPHM, will enable PHM practitioners to easily source, download, and preprocess publicly available PHM datasets in only a few lines of code. Researchers can use the preprocessed data from PyPHM, for example, for feature engineering or machine learning experiments.
Figure 6 illustrates the class hierarchy of the PyPHM software package. Specific datasets are accessible by their own class (beneath the base PyPHM class). The dataset class allows the downloading and extraction of the dataset, along with general preprocessing methods. Finally, simple data preprocessing methods, such as windowing, are constructed in their own preprocessing method classes. PyPHM is implemented in Python and relies on common open-source libraries like NumPy [Harris \BOthers. (\APACyear2020)] and SciPy [Virtanen \BOthers. (\APACyear2020)].
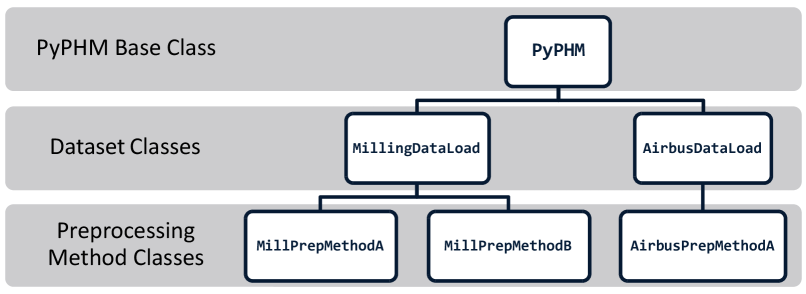
Currently, there are three datasets implemented in PyPHM: the UC-Berkeley Milling Dataset [Agogino \BBA Goebel (\APACyear2007)], the IMS Bearing Dataset [Lee \BOthers. (\APACyear2007)], and the Airbus Helicopter Accelerometer Dataset [Garcia \BOthers. (\APACyear2021)]. More will be implemented in the future.
PyPHM seeks to be a domain specific resource, within PHM, that can assist researchers in conducting reproducible computational research. We highlight three challenges, as noted in Section 5, that PyPHM seeks to specifically addresses.
-
1.
Challenge of expertise: PyPHM abstracts away the complexity of downloading, manipulating, and preprocessing PHM datasets. Thus, a broader audience can engage with PHM datasets, and do so in a way that can be readily reproduced by others. In addition, PyPHM is built upon common open-source tooling. Individuals can educate themselves on these tools, from a PHM context, through PyPHM’s documentation and examples.
-
2.
Challenge of extra effort: PyPHM allows individuals to quickly implement a standardized workflow that other researchers have used. This saves time, reduces effort, and prevents the implementation of ad-hoc workflows that are difficult for others to reproduce.
-
3.
Challenge due to lack of data: Currently, PHM datasets are dispersed across the internet. PyPHM can act as an index for these PHM datasets and a central location to access them. PyPHM can also help individuals share and explore under-utilized PHM datasets once more datasets are added.
PyPHM is in active development. We welcome feedback and contributions to this nascent open-source software project.
7 Conclusion
Computation is used across the breadth of science, and certainly within PHM. However, creating reproducible computational research remains a challenge due to the expertise required, motivational challenges, and lack of domain specific resources. Our survey of more than 300 articles, from publications engaged in PHM research, demonstrates that most researchers within PHM do not provide access to their data or code.
Despite these challenges, there are clear motivations and opportunities for improving reproducible computational research. In this work, we have highlighted three of the personal benefits of conducting reproducible computational research. Namely, creating reproducible computational research can increase a researcher’s exposure, improve career opportunities, and increase one’s sense of satisfaction.
Furthermore, we have identified a need for an open-source software package to assist PHM researchers in accessing and preprocessing common PHM datasets. As such, we have created PyPHM, and we encourage others to assist in our efforts, either through contributions or suggestions.
References
- Agogino \BBA Goebel (\APACyear2007) \APACinsertmetastaragogino2007milling{APACrefauthors}Agogino, A.\BCBT \BBA Goebel, K. \APACrefYearMonthDay2007. \BBOQ\APACrefatitleMilling data set. NASA Ames Prognostics Data Repository Milling data set. nasa ames prognostics data repository.\BBCQ \APACjournalVolNumPagesMoffett Field, CA. {APACrefURL} https://ti.arc.nasa.gov/tech/dash/groups/pcoe/prognostic-data-repository/ \PrintBackRefs\CurrentBib
- Astfalck \BOthers. (\APACyear2016) \APACinsertmetastarastfalck2016modelling{APACrefauthors}Astfalck, L., Hodkiewicz, M., Keating, A., Cripps, E.\BCBL \BBA Pecht, M. \APACrefYearMonthDay2016. \BBOQ\APACrefatitleA modelling ecosystem for prognostics A modelling ecosystem for prognostics.\BBCQ \BIn \APACrefbtitleAnnual Conference of the PHM Society Annual conference of the phm society (\BVOL 8). \PrintBackRefs\CurrentBib
- Bird \BOthers. (\APACyear2009) \APACinsertmetastarBird_Natural_Language_Processing_2009{APACrefauthors}Bird, S., Klein, E.\BCBL \BBA Loper, E. \APACrefYear2009. \APACrefbtitleNatural Language Processing with Python: Analyzing Text with the Natural Language Toolkit Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit. \APACaddressPublisherO’Reilly Media, Inc. \PrintBackRefs\CurrentBib
- Bitzer \BOthers. (\APACyear2007) \APACinsertmetastarbitzer2007intrinsic{APACrefauthors}Bitzer, J., Schrettl, W.\BCBL \BBA Schröder, P\BPBIJ. \APACrefYearMonthDay2007. \BBOQ\APACrefatitleIntrinsic motivation in open source software development Intrinsic motivation in open source software development.\BBCQ \APACjournalVolNumPagesJournal of comparative economics351160–169. \PrintBackRefs\CurrentBib
- Buckheit \BBA Donoho (\APACyear1995) \APACinsertmetastarbuckheit1995wavelab{APACrefauthors}Buckheit, J\BPBIB.\BCBT \BBA Donoho, D\BPBIL. \APACrefYearMonthDay1995. \BBOQ\APACrefatitleWavelab and reproducible research Wavelab and reproducible research.\BBCQ \BIn \APACrefbtitleWavelets and statistics Wavelets and statistics (\BPGS 55–81). \APACaddressPublisherSpringer. \PrintBackRefs\CurrentBib
- Chollet (\APACyear2021) \APACinsertmetastarchollet2021deep{APACrefauthors}Chollet, F. \APACrefYear2021. \APACrefbtitleDeep learning with Python Deep learning with python. \APACaddressPublisherSimon and Schuster. \PrintBackRefs\CurrentBib
- Christensen \BOthers. (\APACyear2019) \APACinsertmetastarchristensen2019study{APACrefauthors}Christensen, G., Dafoe, A., Miguel, E., Moore, D\BPBIA.\BCBL \BBA Rose, A\BPBIK. \APACrefYearMonthDay2019. \BBOQ\APACrefatitleA study of the impact of data sharing on article citations using journal policies as a natural experiment A study of the impact of data sharing on article citations using journal policies as a natural experiment.\BBCQ \APACjournalVolNumPagesPLoS One1412e0225883. \PrintBackRefs\CurrentBib
- Colavizza \BOthers. (\APACyear2020) \APACinsertmetastarcolavizza2020citation{APACrefauthors}Colavizza, G., Hrynaszkiewicz, I., Staden, I., Whitaker, K.\BCBL \BBA McGillivray, B. \APACrefYearMonthDay2020. \BBOQ\APACrefatitleThe citation advantage of linking publications to research data The citation advantage of linking publications to research data.\BBCQ \APACjournalVolNumPagesPloS one154e0230416. \PrintBackRefs\CurrentBib
- Donoho \BOthers. (\APACyear2008) \APACinsertmetastardonoho2008reproducible{APACrefauthors}Donoho, D\BPBIL., Maleki, A., Rahman, I\BPBIU., Shahram, M.\BCBL \BBA Stodden, V. \APACrefYearMonthDay2008. \BBOQ\APACrefatitleReproducible research in computational harmonic analysis Reproducible research in computational harmonic analysis.\BBCQ \APACjournalVolNumPagesComputing in Science & Engineering1118–18. \PrintBackRefs\CurrentBib
- Dorch \BOthers. (\APACyear2015) \APACinsertmetastardorch2015data{APACrefauthors}Dorch, B\BPBIF., Drachen, T\BPBIM.\BCBL \BBA Ellegaard, O. \APACrefYearMonthDay2015. \BBOQ\APACrefatitleThe data sharing advantage in astrophysics The data sharing advantage in astrophysics.\BBCQ \APACjournalVolNumPagesProceedings of the International Astronomical Union11A29A172–175. \PrintBackRefs\CurrentBib
- Esteban \BOthers. (\APACyear2019) \APACinsertmetastaresteban2019fmriprep{APACrefauthors}Esteban, O., Markiewicz, C\BPBIJ., Blair, R\BPBIW., Moodie, C\BPBIA., Isik, A\BPBII., Erramuzpe, A.\BDBLothers \APACrefYearMonthDay2019. \BBOQ\APACrefatitlefMRIPrep: a robust preprocessing pipeline for functional MRI fmriprep: a robust preprocessing pipeline for functional mri.\BBCQ \APACjournalVolNumPagesNature methods161111–116. \PrintBackRefs\CurrentBib
- Frachtenberg (\APACyear2022) \APACinsertmetastarfrachtenberg2022research{APACrefauthors}Frachtenberg, E. \APACrefYearMonthDay2022. \BBOQ\APACrefatitleResearch artifacts and citations in computer systems papers Research artifacts and citations in computer systems papers.\BBCQ \APACjournalVolNumPagesPeerJ Computer Science8e887. \PrintBackRefs\CurrentBib
- Fu \BBA Hughey (\APACyear2019) \APACinsertmetastarfu2019meta{APACrefauthors}Fu, D\BPBIY.\BCBT \BBA Hughey, J\BPBIJ. \APACrefYearMonthDay2019. \BBOQ\APACrefatitleMeta-Research: Releasing a preprint is associated with more attention and citations for the peer-reviewed article Meta-research: Releasing a preprint is associated with more attention and citations for the peer-reviewed article.\BBCQ \APACjournalVolNumPagesElife8e52646. \PrintBackRefs\CurrentBib
- Garcia \BOthers. (\APACyear2021) \APACinsertmetastargarcia2021temporal{APACrefauthors}Garcia, G\BPBIR., Michau, G., Ducoffe, M., Gupta, J\BPBIS.\BCBL \BBA Fink, O. \APACrefYearMonthDay2021. \BBOQ\APACrefatitleTemporal signals to images: Monitoring the condition of industrial assets with deep learning image processing algorithms Temporal signals to images: Monitoring the condition of industrial assets with deep learning image processing algorithms.\BBCQ \APACjournalVolNumPagesProceedings of the Institution of Mechanical Engineers, Part O: Journal of Risk and Reliability1748006X21994446. \PrintBackRefs\CurrentBib
- Gundersen \BOthers. (\APACyear2018) \APACinsertmetastargundersen2018reproducible{APACrefauthors}Gundersen, O\BPBIE., Gil, Y.\BCBL \BBA Aha, D\BPBIW. \APACrefYearMonthDay2018. \BBOQ\APACrefatitleOn reproducible AI: Towards reproducible research, open science, and digital scholarship in AI publications On reproducible ai: Towards reproducible research, open science, and digital scholarship in ai publications.\BBCQ \APACjournalVolNumPagesAI magazine39356–68. \PrintBackRefs\CurrentBib
- Harris \BOthers. (\APACyear2020) \APACinsertmetastarharris2020array{APACrefauthors}Harris, C\BPBIR., Millman, K\BPBIJ., van der Walt, S\BPBIJ., Gommers, R., Virtanen, P., Cournapeau, D.\BDBLOliphant, T\BPBIE. \APACrefYearMonthDay2020\APACmonth09. \BBOQ\APACrefatitleArray programming with NumPy Array programming with NumPy.\BBCQ \APACjournalVolNumPagesNature5857825357–362. {APACrefURL} https://doi.org/10.1038/s41586-020-2649-2 {APACrefDOI} 10.1038/s41586-020-2649-2 \PrintBackRefs\CurrentBib
- Hars \BBA Ou (\APACyear\bibnodate) \APACinsertmetastarhars34working{APACrefauthors}Hars, A.\BCBT \BBA Ou, S. \APACrefYearMonthDay\bibnodate. \BBOQ\APACrefatitleWorking for Free?–Motivations of Participating in Open Source Projects; 2001 Working for free?–motivations of participating in open source projects; 2001.\BBCQ \BIn \APACrefbtitle34th Annual Hawaii International Conference on System Sciences (HICSS-34), Havaí 34th annual hawaii international conference on system sciences (hicss-34), havaí (\BPGS 25–39). \PrintBackRefs\CurrentBib
- Henneken \BBA Accomazzi (\APACyear2011) \APACinsertmetastarhenneken2011linking{APACrefauthors}Henneken, E\BPBIA.\BCBT \BBA Accomazzi, A. \APACrefYearMonthDay2011. \BBOQ\APACrefatitleLinking to data-effect on citation rates in astronomy Linking to data-effect on citation rates in astronomy.\BBCQ \APACjournalVolNumPagesarXiv preprint arXiv:1111.3618. \PrintBackRefs\CurrentBib
- Hu \BOthers. (\APACyear2022) \APACinsertmetastarhu2022prognostics{APACrefauthors}Hu, Y., Miao, X., Si, Y., Pan, E.\BCBL \BBA Zio, E. \APACrefYearMonthDay2022. \BBOQ\APACrefatitlePrognostics and health management: A review from the perspectives of design, development and decision Prognostics and health management: A review from the perspectives of design, development and decision.\BBCQ \APACjournalVolNumPagesReliability Engineering & System Safety217108063. \PrintBackRefs\CurrentBib
- Ince \BOthers. (\APACyear2012) \APACinsertmetastarince2012case{APACrefauthors}Ince, D\BPBIC., Hatton, L.\BCBL \BBA Graham-Cumming, J. \APACrefYearMonthDay2012. \BBOQ\APACrefatitleThe case for open computer programs The case for open computer programs.\BBCQ \APACjournalVolNumPagesNature4827386485–488. \PrintBackRefs\CurrentBib
- \APACcitebtitleAn introduction to the foundations of neuro data science (\APACyear\bibnodate) \APACinsertmetastarneurodatascience\APACrefbtitleAn introduction to the foundations of neuro data science. An introduction to the foundations of neuro data science. \APACrefYearMonthDay\bibnodate. \APACaddressPublisherMcGill University. {APACrefURL} https://neurodatascience.github.io/QLS612-Overview/ \PrintBackRefs\CurrentBib
- Lee \BOthers. (\APACyear2007) \APACinsertmetastarlee2007bearing{APACrefauthors}Lee, J., Qiu, H., Yu, G., Lin, J.\BCBL \BOthersPeriod. \APACrefYearMonthDay2007. \BBOQ\APACrefatitleBearing data set Bearing data set.\BBCQ \APACjournalVolNumPagesIMS, University of Cincinnati, NASA Ames Prognostics Data Repository, Rexnord Technical Services. \PrintBackRefs\CurrentBib
- National Academies of Sciences, Engineering, and Medicine and others (\APACyear2019) \APACinsertmetastarnational2019reproducibility{APACrefauthors}National Academies of Sciences, Engineering, and Medicine and others. \APACrefYearMonthDay2019. \BBOQ\APACrefatitleReproducibility and replicability in science Reproducibility and replicability in science.\BBCQ \PrintBackRefs\CurrentBib
- Paszke \BOthers. (\APACyear2019) \APACinsertmetastarNEURIPS2019_9015{APACrefauthors}Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G.\BDBLChintala, S. \APACrefYearMonthDay2019. \BBOQ\APACrefatitlePyTorch: An Imperative Style, High-Performance Deep Learning Library Pytorch: An imperative style, high-performance deep learning library.\BBCQ \BIn H. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. Fox\BCBL \BBA R. Garnett (\BEDS), \APACrefbtitleAdvances in Neural Information Processing Systems 32 Advances in neural information processing systems 32 (\BPGS 8024–8035). \APACaddressPublisherCurran Associates, Inc. {APACrefURL} http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf \PrintBackRefs\CurrentBib
- Perone \BOthers. (\APACyear2018) \APACinsertmetastarmedicaltorchPerone{APACrefauthors}Perone, C\BPBIS., cclauss, Saravia, E., Ballester, P\BPBIL.\BCBL \BBA MohitTare. \APACrefYearMonthDay2018. \BBOQ\APACrefatitlemedicaltorch: An open-source pytorch medical imaging framework medicaltorch: An open-source pytorch medical imaging framework.\BBCQ \APACjournalVolNumPagesGitHub. {APACrefURL} https://doi.org/10.5281/zenodo.1495335 {APACrefDOI} 10.5281/zenodo.1495335 \PrintBackRefs\CurrentBib
- Piwowar \BOthers. (\APACyear2007) \APACinsertmetastarpiwowar2007sharing{APACrefauthors}Piwowar, H\BPBIA., Day, R\BPBIS.\BCBL \BBA Fridsma, D\BPBIB. \APACrefYearMonthDay2007. \BBOQ\APACrefatitleSharing detailed research data is associated with increased citation rate Sharing detailed research data is associated with increased citation rate.\BBCQ \APACjournalVolNumPagesPloS one23e308. \PrintBackRefs\CurrentBib
- Piwowar \BBA Vision (\APACyear2013) \APACinsertmetastarpiwowar2013data{APACrefauthors}Piwowar, H\BPBIA.\BCBT \BBA Vision, T\BPBIJ. \APACrefYearMonthDay2013. \BBOQ\APACrefatitleData reuse and the open data citation advantage Data reuse and the open data citation advantage.\BBCQ \APACjournalVolNumPagesPeerJ1e175. \PrintBackRefs\CurrentBib
- \APACcitebtitlePrinciples, Statistical and Computational Tools for Reproducible Data Science (\APACyear2017) \APACinsertmetastarharvard2017reproducible\APACrefbtitlePrinciples, Statistical and Computational Tools for Reproducible Data Science. Principles, statistical and computational tools for reproducible data science. \APACrefYearMonthDay2017Oct. \APACaddressPublisherHarvard University. {APACrefURL} https://pll.harvard.edu/course/principles-statistical-and-computational-tools-reproducible-data-science \PrintBackRefs\CurrentBib
- Rainie \BBA Anderson (\APACyear2017) \APACinsertmetastarrainie2017future{APACrefauthors}Rainie, L.\BCBT \BBA Anderson, J. \APACrefYearMonthDay2017. \BBOQ\APACrefatitleThe Future of Jobs and Jobs Training. The future of jobs and jobs training.\BBCQ \APACjournalVolNumPagesPew Research Center. \PrintBackRefs\CurrentBib
- \APACcitebtitleReproducible and Collaborative Data Science (\APACyear\bibnodate) \APACinsertmetastarucberkeleyreproducible\APACrefbtitleReproducible and Collaborative Data Science. Reproducible and collaborative data science. \APACrefYearMonthDay\bibnodate. \APACaddressPublisherUniversity of California, Berkeley. {APACrefURL} https://berkeley-stat159-f17.github.io/stat159-f17/ \PrintBackRefs\CurrentBib
- Stodden \BOthers. (\APACyear2013) \APACinsertmetastarstodden2013toward{APACrefauthors}Stodden, V., Guo, P.\BCBL \BBA Ma, Z. \APACrefYearMonthDay2013. \BBOQ\APACrefatitleToward reproducible computational research: an empirical analysis of data and code policy adoption by journals Toward reproducible computational research: an empirical analysis of data and code policy adoption by journals.\BBCQ \APACjournalVolNumPagesPloS one86e67111. \PrintBackRefs\CurrentBib
- Stodden \BOthers. (\APACyear2018) \APACinsertmetastarstodden2018enabling{APACrefauthors}Stodden, V., Krafczyk, M\BPBIS.\BCBL \BBA Bhaskar, A. \APACrefYearMonthDay2018. \BBOQ\APACrefatitleEnabling the verification of computational results: An empirical evaluation of computational reproducibility Enabling the verification of computational results: An empirical evaluation of computational reproducibility.\BBCQ \BIn \APACrefbtitleProceedings of the First International Workshop on Practical Reproducible Evaluation of Computer Systems Proceedings of the first international workshop on practical reproducible evaluation of computer systems (\BPGS 1–5). \PrintBackRefs\CurrentBib
- \APACciteatitleTrouble at the lab (\APACyear2013) \APACinsertmetastartrouble_lab_2013\BBOQ\APACrefatitleTrouble at the lab Trouble at the lab.\BBCQ \APACrefYearMonthDay2013Oct. \APACjournalVolNumPagesThe Economist. {APACrefURL} https://www.economist.com/briefing/2013/10/18/trouble-at-the-lab \PrintBackRefs\CurrentBib
- Vanderplas \BOthers. (\APACyear2012) \APACinsertmetastarastroML{APACrefauthors}Vanderplas, J., Connolly, A., Ivezić, Ž.\BCBL \BBA Gray, A. \APACrefYearMonthDay2012oct.. \BBOQ\APACrefatitleIntroduction to astroML: Machine learning for astrophysics Introduction to astroml: Machine learning for astrophysics.\BBCQ \BIn \APACrefbtitleConference on Intelligent Data Understanding (CIDU) Conference on intelligent data understanding (cidu) (\BPG 47 -54). {APACrefDOI} 10.1109/CIDU.2012.6382200 \PrintBackRefs\CurrentBib
- Virtanen \BOthers. (\APACyear2020) \APACinsertmetastar2020SciPy-NMeth{APACrefauthors}Virtanen, P., Gommers, R., Oliphant, T\BPBIE., Haberland, M., Reddy, T., Cournapeau, D.\BDBLSciPy 1.0 Contributors \APACrefYearMonthDay2020. \BBOQ\APACrefatitleSciPy 1.0: Fundamental Algorithms for Scientific Computing in Python SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python.\BBCQ \APACjournalVolNumPagesNature Methods17261–272. {APACrefDOI} 10.1038/s41592-019-0686-2 \PrintBackRefs\CurrentBib
- Wahlquist \BOthers. (\APACyear2018) \APACinsertmetastarwahlquist2018dissemination{APACrefauthors}Wahlquist, A\BPBIE., Muhammad, L\BPBIN., Herbert, T\BPBIL., Ramakrishnan, V.\BCBL \BBA Nietert, P\BPBIJ. \APACrefYearMonthDay2018. \BBOQ\APACrefatitleDissemination of novel biostatistics methods: Impact of programming code availability and other characteristics on article citations Dissemination of novel biostatistics methods: Impact of programming code availability and other characteristics on article citations.\BBCQ \APACjournalVolNumPagesPloS one138e0201590. \PrintBackRefs\CurrentBib
- Wang \BOthers. (\APACyear2021) \APACinsertmetastarwang2021recent{APACrefauthors}Wang, W., Taylor, J.\BCBL \BBA Rees, R\BPBIJ. \APACrefYearMonthDay2021. \BBOQ\APACrefatitleRecent Advancement of Deep Learning Applications to Machine Condition Monitoring Part 1: A Critical Review Recent advancement of deep learning applications to machine condition monitoring part 1: A critical review.\BBCQ \APACjournalVolNumPagesAcoustics Australia1–13. \PrintBackRefs\CurrentBib
- Wilson (\APACyear2014) \APACinsertmetastarwilson2014software{APACrefauthors}Wilson, G. \APACrefYearMonthDay2014. \BBOQ\APACrefatitleSoftware Carpentry: lessons learned Software carpentry: lessons learned.\BBCQ \APACjournalVolNumPagesF1000Research3. \PrintBackRefs\CurrentBib
- Zhao \BOthers. (\APACyear2019) \APACinsertmetastarzhao2019deep{APACrefauthors}Zhao, R., Yan, R., Chen, Z., Mao, K., Wang, P.\BCBL \BBA Gao, R\BPBIX. \APACrefYearMonthDay2019. \BBOQ\APACrefatitleDeep learning and its applications to machine health monitoring Deep learning and its applications to machine health monitoring.\BBCQ \APACjournalVolNumPagesMechanical Systems and Signal Processing115213–237. \PrintBackRefs\CurrentBib
- Zilberman \BBA Ice (\APACyear2021) \APACinsertmetastarzilberman2021computer{APACrefauthors}Zilberman, A.\BCBT \BBA Ice, L. \APACrefYearMonthDay2021. \BBOQ\APACrefatitleWhy computer occupations are behind strong STEM employment growth in the 2019–29 decade Why computer occupations are behind strong stem employment growth in the 2019–29 decade.\BBCQ \APACjournalVolNumPagesComputer45,164.611–5. \PrintBackRefs\CurrentBib
Appendix


