MIDLMedical Imaging with Deep Learning
\jmlrpages
\jmlryear2019
\jmlrworkshopMIDL 2019 – Extended Abstract Track
\midlauthor\NameDavid Zimmerer\nametag1
\NameSimon Kohl\nametag1
\NameJens Petersen\nametag1
\NameFabian Isensee\nametag1
\NameKlaus Maier-Hein\nametag1
\addr1 German Cancer Research Center (DKFZ), Heidelberg, Germany
Context-encoding Variational Autoencoder
for Unsupervised Anomaly Detection
Abstract
Unsupervised learning can leverage large-scale data sources without the need for annotations. In this context, deep learning-based autoencoders have shown great potential in detecting anomalies in medical images. However, especially Variational Autoencoders (VAEs) often fail to capture the high-level structure in the data. We address these shortcomings by proposing the context-encoding Variational Autoencoder (ceVAE), which improves both, the sample- as well as pixelwise results. In our experiments on the BraTS-2017 and ISLES-2015 segmentation benchmarks the ceVAE achieves unsupervised AUROCs of 0.95 and 0.89, respectively, thus outperforming other reported deep-learning based approaches.
1 Introduction
In the last years several computer-aided diagnosis systems have reported near human-level performance [Liu et al.(2017)Liu, Gadepalli, Norouzi, Dahl, Kohlberger, Venugopalan, Boyko, Timofeev, Nelson, Corrado, Hipp, Peng, and Stumpe, Gulshan et al.(2016)Gulshan, Peng, Coram, Stumpe, Wu, Narayanaswamy, Venugopalan, Widner, Madams, Cuadros, and others]. However those approaches are only applicable in a narrow range of cases, where larger amounts of annotated training data is available. Unsupervised approaches on the other hand do not need any annotated data and thus allow broader applicability and independence of human errors in the annotation. Recently, several deep-learning based pixelwise anomaly detection approaches have shown great promise [Abati et al.(2018)Abati, Porrello, Calderara, and Cucchiara, Baur et al.(2018)Baur, Wiestler, Albarqouni, and Navab, Chen et al.(2018)Chen, Pawlowski, Rajchl, Glocker, and Konukoglu, Chen and Konukoglu(2018), Pawlowski et al.(2018)Pawlowski, Lee, Rajchl, McDonagh, Ferrante, Kamnitsas, Cooke, Stevenson, Khetani, Newman, Zeiler, Digby, Coles, Rueckert, Menon, Newcombe, and Glocker, Schlegl et al.(2017)Schlegl, Seeböck, Waldstein, Schmidt-Erfurth, and Langs]. Most of these approaches are built on the reconstruction error of generative models, Variational Autoencoders (VAEs) in particular. However recent research has critiqued VAEs for their low capability to extract high-level structure in the data, and suggested improvements over the base model [Nalisnick et al.(2018)Nalisnick, Matsukawa, Teh, Gorur, and Lakshminarayanan, Zhao et al.(2017)Zhao, Song, and Ermon, Maaløe et al.(2019)Maaløe, Fraccaro, Liévin, and Winther]. Here we use context-encoding [Pathak et al.(2016)Pathak, Krähenbühl, Donahue, Darrell, and Efros] to steer the VAE towards learning more discriminative features. Our results suggest that these features can improve out-of-distribution/anomaly-detection tasks and as such aid VAEs in capturing the data distribution.
2 Methods
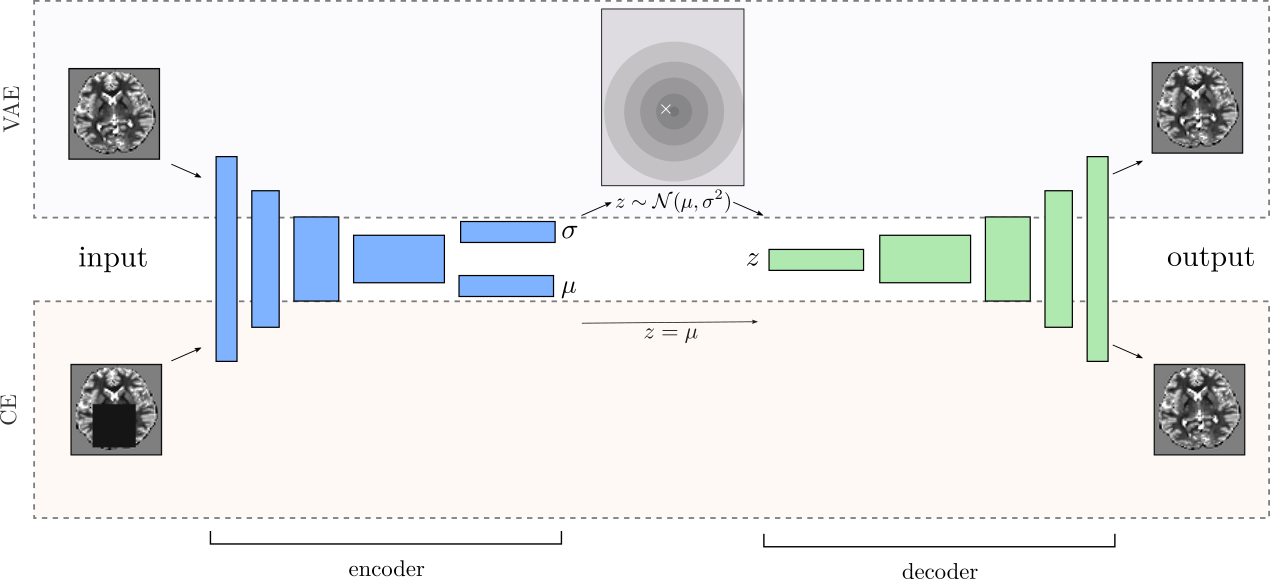
VAE-based methods are, next to flow-based and autoregessive models, one of the most commonly used models for density estimation and out-of-distribution/anomaly-detection tasks. Denoising autoencoders and especially context-encoders (CE) on the other hand were shown to learn discriminative features, invariant to perturbations of the data [Pathak et al.(2016)Pathak, Krähenbühl, Donahue, Darrell, and Efros, Vincent et al.(2010)Vincent, Larochelle, Lajoie, Bengio, and Manzagol]. We propose to combine them into a context-encoding VAE (ceVAE), as shown in Fig. 1a, to learn more discriminative features and to prevent posterior collapse in VAEs. Each data sample is used to train a normal VAE. At the same time, CE-noise perturbed data samples are used to train a CE, where we do not sample from the latent posterior distribution but use the mean as encoding . This results in the following objective:
| (1) |
with neural network encoders , and decoder . weighs the VAE objective against the CE objective (default ).
To detect samplewise anomalies we use the approximated evidence lower bound (ELBO) from the VAE as proxy for the data likelihood and consequently as anomaly score. Since a low ELBO can be either caused by a high reconstruction error or a high KL-divergence , we use both for a pixelwise anomaly score. As commonly done [Baur et al.(2018)Baur, Wiestler, Albarqouni, and Navab, Chen et al.(2018)Chen, Pawlowski, Rajchl, Glocker, and Konukoglu, Pawlowski et al.(2018)Pawlowski, Lee, Rajchl, McDonagh, Ferrante, Kamnitsas, Cooke, Stevenson, Khetani, Newman, Zeiler, Digby, Coles, Rueckert, Menon, Newcombe, and Glocker], we use the reconstruction error directly as pixelwise indication for anomalies. To complement this with a pixelwise anomaly score from the KL-divergence, we use the derivative of the KL-divergence with respect to the input , which was previously reported to show good results on its own [Zimmerer et al.(2018)Zimmerer, Petersen, Kohl, and Maier-Hein]. We combine the two pixelwise scores using pixelwise multiplication:
| (2) |
3 Experiments & Results
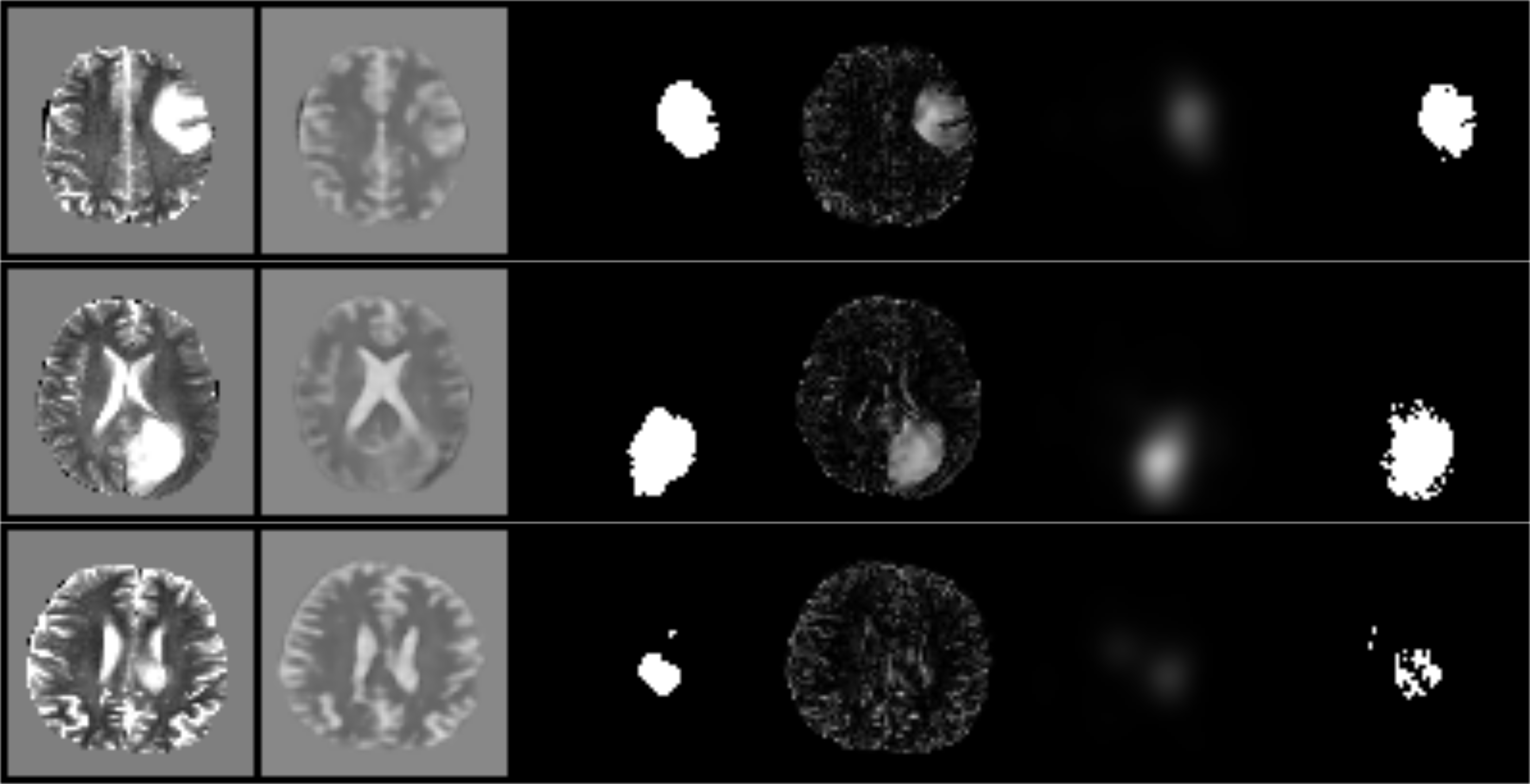
We trained the model on 2D slices of T2-weighted images from the HCP dataset [Van Essen et al.(2012)Van Essen, Ugurbil, Auerbach, Barch, Behrens, Bucholz, Chang, Chen, Corbetta, Curtiss, Della Penna, Feinberg, Glasser, Harel, Heath, Larson-Prior, Marcus, Michalareas, Moeller, Oostenveld, Petersen, Prior, Schlaggar, Smith, Snyder, Xu, Yacoub, and WU-Minn HCP Consortium] (N=1092) to learn the distribution of healthy patient images. The model was tested to find anomalies in the BraTS2017 [Bakas et al.(2017)Bakas, Akbari, Sotiras, Bilello, Rozycki, Kirby, Freymann, Farahani, and Davatzikos, Menze et al.(2015)Menze, Jakab, Bauer, Kalpathy-Cramer, Farahani, Kirby, Burren, Porz, Slotboom, Wiest, Lanczi, Gerstner, Weber, Arbel, Avants, Ayache, Buendia, Collins, Cordier, Corso, Criminisi, Das, Delingette, Demiralp, Durst, Dojat, Doyle, Festa, Forbes, Geremia, Glocker, Golland, Guo, Hamamci, Iftekharuddin, Jena, John, Konukoglu, Lashkari, Mariz, Meier, Pereira, Precup, Price, Raviv, Reza, Ryan, Sarikaya, Schwartz, Shin, Shotton, Silva, Sousa, Subbanna, Szekely, Taylor, Thomas, Tustison, Unal, Vasseur, Wintermark, Ye, Zhao, Zhao, Zikic, Prastawa, Reyes, and Van Leemput] (N=266) and the ISLES2015 [Maier et al.(2017)Maier, Menze, von der Gablentz, Hani, Heinrich, Liebrand, Winzeck, Basit, Bentley, Chen, Christiaens, Dutil, Egger, Feng, Glocker, Götz, Haeck, Halme, Havaei, Iftekharuddin, Jodoin, Kamnitsas, Kellner, Korvenoja, Larochelle, Ledig, Lee, Maes, Mahmood, Maier-Hein, McKinley, Muschelli, Pal, Pei, Rangarajan, Reza, Robben, Rueckert, Salli, Suetens, Wang, Wilms, Kirschke, Kr Amer, Münte, Schramm, Wiest, Handels, and Reyes] (N=20) dataset. For a samplewise detection slices without any annotations were regarded as normal and samples with annotated pixels were regarded as anomalies. For the pixelwise detection, annotated pixels were regarded as anomalies, while pixels without any annotations were regarded as normal. For the encoder and decoder networks, we chose fully convolutional networks with five 2D-Conv-Layers and 2D-Transposed-Conv-Layers respectively with kernel size 4 and stride 2, each layer followed by a LeakyReLU non-linearity. The models were trained with Adam with a learning rate of and a batch size of 64 for 60 epochs.
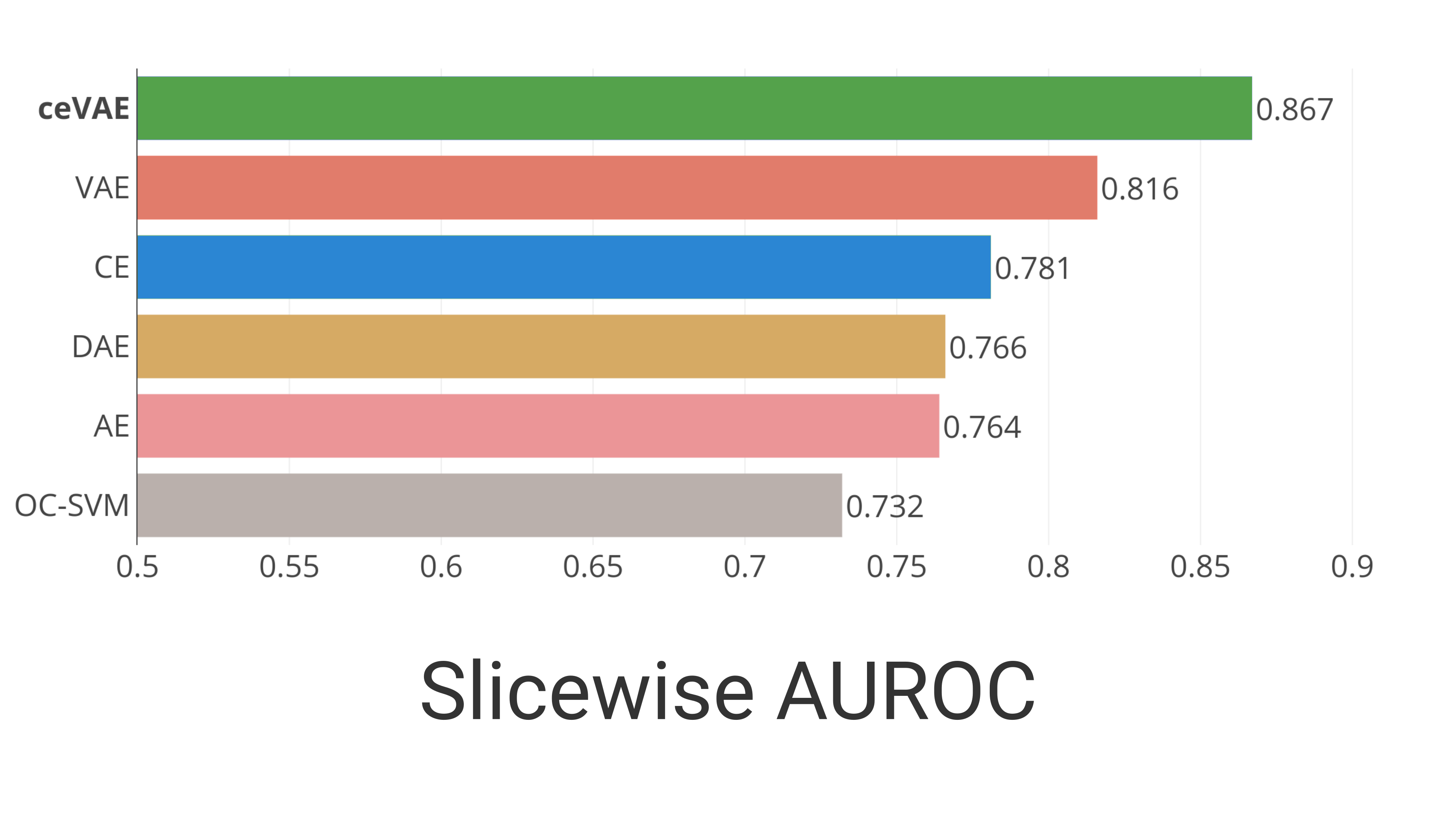
The samplewise results can be seen in Fig. 2a, indicating that the CE-objective can aid VAEs for anomaly detection. To analyze the posterior collapse we inspect the 0.95 quantile of the KL-divergence over the test samples after training. For the ceVAE the 0.95 quantile is 2.93 while for the VAE it is 0.53, indicating that more latent variables are used in the ceVAE and fewer have collapsed.
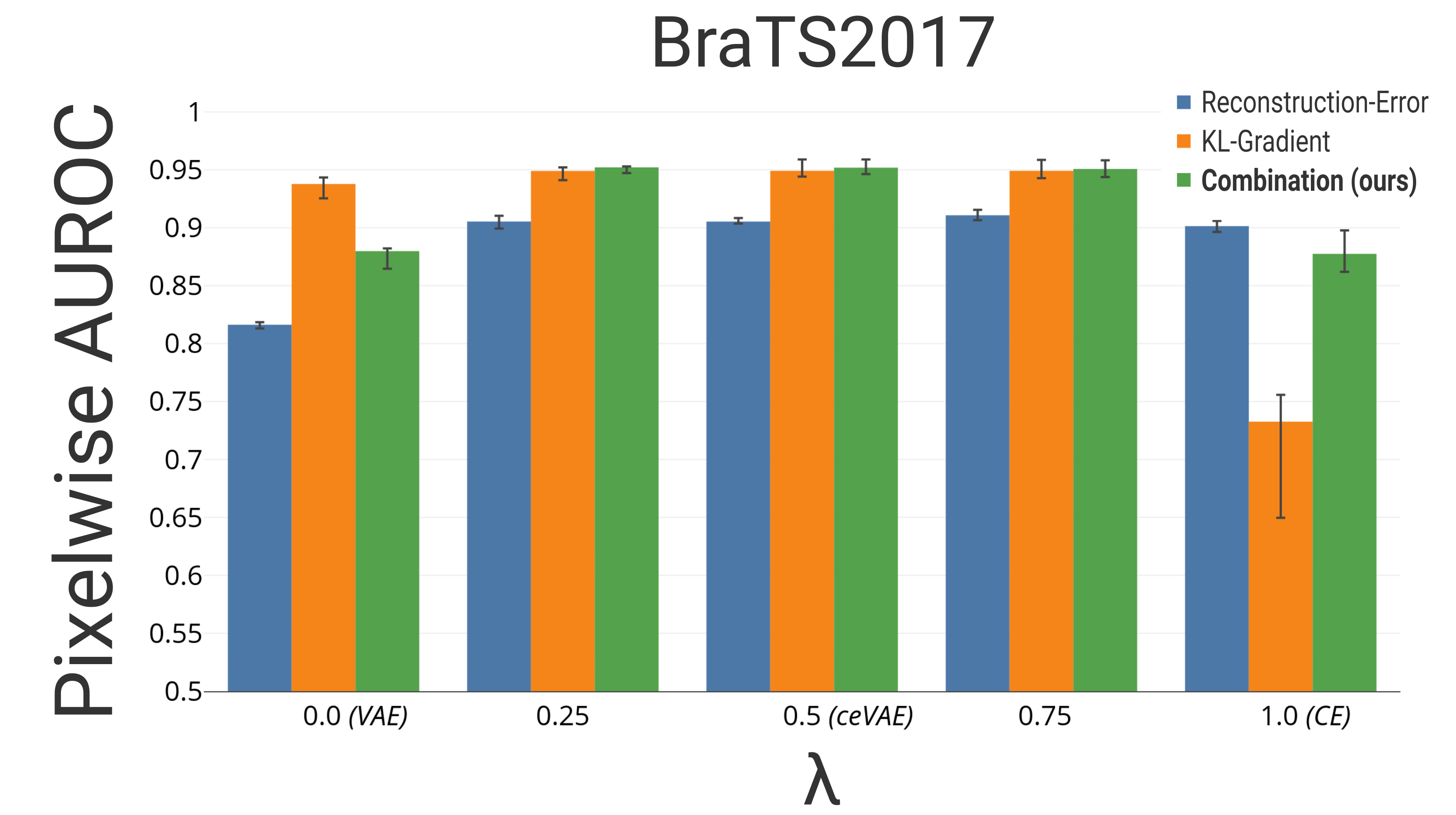
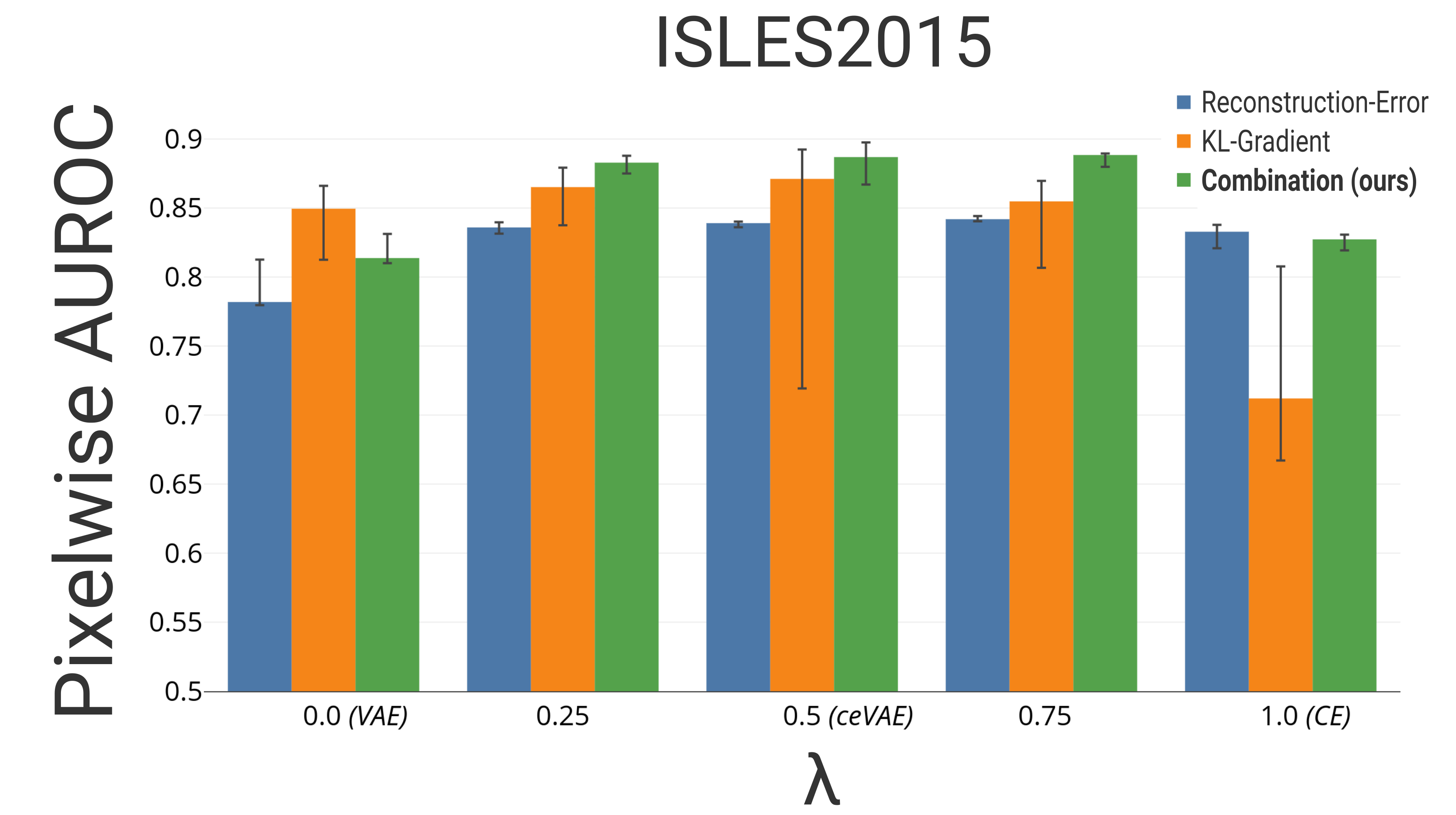
The pixelwise results can be seen in Fig. 2b,c. The combined approach in Eq. 2 as well as its two components are presented individually. Including the CE objective into the VAEs appears to already improve the pixelwise reconstruction-only based detection. The same can also be observed for the KL-Gradient based detection. The best result on the BraTS and ISLES dataset are achieved using for the model proposed in Eq. 1. Using of the BraTS2017 testset to determine a threshold, and calculating the Dice score of the other of the testset, results in a Dice score of 0.52 outperforming previously reported deep-learning based anomaly detection results on similar datasets [Chen et al.(2018)Chen, Pawlowski, Rajchl, Glocker, and Konukoglu]. While the analysis was performed on -sized images, early results with images sizes of show a better performance, in particular on the ISLES2015 dataset with an AUROC of .
4 Discussion & Conclusion
We presented a way to integrate context encoding into VAEs. This leads to a better utilization of the latent-variables and shows better performance detecting out-of-distribution samples/anomalies samplewise and pixelwise. We are confident that this represents an important step towards anomaly detection without the need for annotated data and thus make computer-aided diagnosis applicable to a wider variety of cases.
References
- [Abati et al.(2018)Abati, Porrello, Calderara, and Cucchiara] Davide Abati, Angelo Porrello, Simone Calderara, and Rita Cucchiara. AND: Autoregressive Novelty Detectors. CoRR, abs/1807.01653, 2018.
- [Bakas et al.(2017)Bakas, Akbari, Sotiras, Bilello, Rozycki, Kirby, Freymann, Farahani, and Davatzikos] Spyridon Bakas, Hamed Akbari, Aristeidis Sotiras, Michel Bilello, Martin Rozycki, Justin S. Kirby, John B. Freymann, Keyvan Farahani, and Christos Davatzikos. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data, 4:170117, 2017. ISSN 2052-4463. 10.1038/sdata.2017.117.
- [Baur et al.(2018)Baur, Wiestler, Albarqouni, and Navab] Christoph Baur, Benedikt Wiestler, Shadi Albarqouni, and Nassir Navab. Deep Autoencoding Models for Unsupervised Anomaly Segmentation in Brain MR Images. CoRR, abs/1804.04488, 2018.
- [Chen and Konukoglu(2018)] Xiaoran Chen and Ender Konukoglu. Unsupervised Detection of Lesions in Brain MRI using constrained adversarial auto-encoders. CoRR, abs/1806.04972, 2018.
- [Chen et al.(2018)Chen, Pawlowski, Rajchl, Glocker, and Konukoglu] Xiaoran Chen, Nick Pawlowski, Martin Rajchl, Ben Glocker, and Ender Konukoglu. Deep Generative Models in the Real-World: An Open Challenge from Medical Imaging. CoRR, abs/1806.05452, 2018.
- [Gulshan et al.(2016)Gulshan, Peng, Coram, Stumpe, Wu, Narayanaswamy, Venugopalan, Widner, Madams, Cuadros, and others] Varun Gulshan, Lily Peng, Marc Coram, Martin C Stumpe, Derek Wu, Arunachalam Narayanaswamy, Subhashini Venugopalan, Kasumi Widner, Tom Madams, Jorge Cuadros, and others. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama, 316(22):2402–2410, 2016.
- [Liu et al.(2017)Liu, Gadepalli, Norouzi, Dahl, Kohlberger, Venugopalan, Boyko, Timofeev, Nelson, Corrado, Hipp, Peng, and Stumpe] Yun Liu, Krishna Kumar Gadepalli, Mohammad Norouzi, George Dahl, Timo Kohlberger, Subhashini Venugopalan, Aleksey S. Boyko, Aleksei Timofeev, Philip Q. Nelson, Greg Corrado, Jason Hipp, Lily Peng, and Martin Stumpe. Detecting Cancer Metastases on Gigapixel Pathology Images. Technical report, arXiv, 2017. URL https://arxiv.org/abs/1703.02442.
- [Maaløe et al.(2019)Maaløe, Fraccaro, Liévin, and Winther] Lars Maaløe, Marco Fraccaro, Valentin Liévin, and Ole Winther. BIVA: A Very Deep Hierarchy of Latent Variables for Generative Modeling. arXiv:1902.02102 [cs, stat], February 2019. URL http://arxiv.org/abs/1902.02102. arXiv: 1902.02102.
- [Maier et al.(2017)Maier, Menze, von der Gablentz, Hani, Heinrich, Liebrand, Winzeck, Basit, Bentley, Chen, Christiaens, Dutil, Egger, Feng, Glocker, Götz, Haeck, Halme, Havaei, Iftekharuddin, Jodoin, Kamnitsas, Kellner, Korvenoja, Larochelle, Ledig, Lee, Maes, Mahmood, Maier-Hein, McKinley, Muschelli, Pal, Pei, Rangarajan, Reza, Robben, Rueckert, Salli, Suetens, Wang, Wilms, Kirschke, Kr Amer, Münte, Schramm, Wiest, Handels, and Reyes] Oskar Maier, Bjoern H. Menze, Janina von der Gablentz, Levin Hani, Mattias P. Heinrich, Matthias Liebrand, Stefan Winzeck, Abdul Basit, Paul Bentley, Liang Chen, Daan Christiaens, Francis Dutil, Karl Egger, Chaolu Feng, Ben Glocker, Michael Götz, Tom Haeck, Hanna-Leena Halme, Mohammad Havaei, Khan M. Iftekharuddin, Pierre-Marc Jodoin, Konstantinos Kamnitsas, Elias Kellner, Antti Korvenoja, Hugo Larochelle, Christian Ledig, Jia-Hong Lee, Frederik Maes, Qaiser Mahmood, Klaus H. Maier-Hein, Richard McKinley, John Muschelli, Chris Pal, Linmin Pei, Janaki Raman Rangarajan, Syed M. S. Reza, David Robben, Daniel Rueckert, Eero Salli, Paul Suetens, Ching-Wei Wang, Matthias Wilms, Jan S. Kirschke, Ulrike M. Kr Amer, Thomas F. Münte, Peter Schramm, Roland Wiest, Heinz Handels, and Mauricio Reyes. ISLES 2015 - A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Medical Image Analysis, 35:250–269, 2017. ISSN 1361-8423. 10.1016/j.media.2016.07.009.
- [Menze et al.(2015)Menze, Jakab, Bauer, Kalpathy-Cramer, Farahani, Kirby, Burren, Porz, Slotboom, Wiest, Lanczi, Gerstner, Weber, Arbel, Avants, Ayache, Buendia, Collins, Cordier, Corso, Criminisi, Das, Delingette, Demiralp, Durst, Dojat, Doyle, Festa, Forbes, Geremia, Glocker, Golland, Guo, Hamamci, Iftekharuddin, Jena, John, Konukoglu, Lashkari, Mariz, Meier, Pereira, Precup, Price, Raviv, Reza, Ryan, Sarikaya, Schwartz, Shin, Shotton, Silva, Sousa, Subbanna, Szekely, Taylor, Thomas, Tustison, Unal, Vasseur, Wintermark, Ye, Zhao, Zhao, Zikic, Prastawa, Reyes, and Van Leemput] Bjoern H. Menze, Andras Jakab, Stefan Bauer, Jayashree Kalpathy-Cramer, Keyvan Farahani, Justin Kirby, Yuliya Burren, Nicole Porz, Johannes Slotboom, Roland Wiest, Levente Lanczi, Elizabeth Gerstner, Marc-André Weber, Tal Arbel, Brian B. Avants, Nicholas Ayache, Patricia Buendia, D. Louis Collins, Nicolas Cordier, Jason J. Corso, Antonio Criminisi, Tilak Das, Hervé Delingette, Cagatay Demiralp, Christopher R. Durst, Michel Dojat, Senan Doyle, Joana Festa, Florence Forbes, Ezequiel Geremia, Ben Glocker, Polina Golland, Xiaotao Guo, Andac Hamamci, Khan M. Iftekharuddin, Raj Jena, Nigel M. John, Ender Konukoglu, Danial Lashkari, José Antonió Mariz, Raphael Meier, Sérgio Pereira, Doina Precup, Stephen J. Price, Tammy Riklin Raviv, Syed M. S. Reza, Michael Ryan, Duygu Sarikaya, Lawrence Schwartz, Hoo-Chang Shin, Jamie Shotton, Carlos A. Silva, Nuno Sousa, Nagesh K. Subbanna, Gabor Szekely, Thomas J. Taylor, Owen M. Thomas, Nicholas J. Tustison, Gozde Unal, Flor Vasseur, Max Wintermark, Dong Hye Ye, Liang Zhao, Binsheng Zhao, Darko Zikic, Marcel Prastawa, Mauricio Reyes, and Koen Van Leemput. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE transactions on medical imaging, 34(10):1993–2024, October 2015. ISSN 1558-254X. 10.1109/TMI.2014.2377694.
- [Nalisnick et al.(2018)Nalisnick, Matsukawa, Teh, Gorur, and Lakshminarayanan] Eric Nalisnick, Akihiro Matsukawa, Yee Whye Teh, Dilan Gorur, and Balaji Lakshminarayanan. Do Deep Generative Models Know What They Don’t Know? arXiv:1810.09136 [cs, stat], October 2018. URL http://arxiv.org/abs/1810.09136. arXiv: 1810.09136.
- [Pathak et al.(2016)Pathak, Krähenbühl, Donahue, Darrell, and Efros] Deepak Pathak, Philipp Krähenbühl, Jeff Donahue, Trevor Darrell, and Alexei A. Efros. Context Encoders: Feature Learning by Inpainting. CVPR, pages 2536–2544, 2016.
- [Pawlowski et al.(2018)Pawlowski, Lee, Rajchl, McDonagh, Ferrante, Kamnitsas, Cooke, Stevenson, Khetani, Newman, Zeiler, Digby, Coles, Rueckert, Menon, Newcombe, and Glocker] Nick Pawlowski, Matthew C. H. Lee, Martin Rajchl, Steven McDonagh, Enzo Ferrante, Konstantinos Kamnitsas, Sam Cooke, Susan K. Stevenson, Aneesh M. Khetani, Tom Newman, Fred A. Zeiler, Richard John Digby, Jonathan P. Coles, Daniel Rueckert, David K. Menon, Virginia F. J. Newcombe, and Ben Glocker. Unsupervised Lesion Detection in Brain CT using Bayesian Convolutional Autoencoders. 2018.
- [Schlegl et al.(2017)Schlegl, Seeböck, Waldstein, Schmidt-Erfurth, and Langs] Thomas Schlegl, Philipp Seeböck, Sebastian M. Waldstein, Ursula Schmidt-Erfurth, and Georg Langs. Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery. In Marc Niethammer, Martin Styner, Stephen Aylward, Hongtu Zhu, Ipek Oguz, Pew-Thian Yap, and Dinggang Shen, editors, IPMI, Lecture Notes in Computer Science, pages 146–157. Springer, 2017. ISBN 978-3-319-59050-9.
- [Van Essen et al.(2012)Van Essen, Ugurbil, Auerbach, Barch, Behrens, Bucholz, Chang, Chen, Corbetta, Curtiss, Della Penna, Feinberg, Glasser, Harel, Heath, Larson-Prior, Marcus, Michalareas, Moeller, Oostenveld, Petersen, Prior, Schlaggar, Smith, Snyder, Xu, Yacoub, and WU-Minn HCP Consortium] D. C. Van Essen, K. Ugurbil, E. Auerbach, D. Barch, T. E. J. Behrens, R. Bucholz, A. Chang, L. Chen, M. Corbetta, S. W. Curtiss, S. Della Penna, D. Feinberg, M. F. Glasser, N. Harel, A. C. Heath, L. Larson-Prior, D. Marcus, G. Michalareas, S. Moeller, R. Oostenveld, S. E. Petersen, F. Prior, B. L. Schlaggar, S. M. Smith, A. Z. Snyder, J. Xu, E. Yacoub, and WU-Minn HCP Consortium. The Human Connectome Project: a data acquisition perspective. NeuroImage, 62(4):2222–2231, October 2012. ISSN 1095-9572. 10.1016/j.neuroimage.2012.02.018.
- [Vincent et al.(2010)Vincent, Larochelle, Lajoie, Bengio, and Manzagol] Pascal Vincent, Hugo Larochelle, Isabelle Lajoie, Yoshua Bengio, and Pierre-Antoine Manzagol. Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion. JMLR, 11(Dec):3371–3408, 2010. ISSN ISSN 1533-7928. URL http://www.jmlr.org/papers/v11/vincent10a.html.
- [Zhao et al.(2017)Zhao, Song, and Ermon] Shengjia Zhao, Jiaming Song, and Stefano Ermon. Learning Hierarchical Features from Generative Models. February 2017. URL http://arxiv.org/abs/1702.08396.
- [Zimmerer et al.(2018)Zimmerer, Petersen, Kohl, and Maier-Hein] David Zimmerer, Jens Petersen, Simon AA Kohl, and Klaus H Maier-Hein. A Case for the Score: Identifying Image Anomalies using Variational Autoencoder Gradients. 2018.