Few-Shot Cross-lingual Transfer for Coarse-grained
De-identification of Code-Mixed Clinical Texts
Abstract
Despite the advances in digital healthcare systems offering curated structured knowledge, much of the critical information still lies in large volumes of unlabeled and unstructured clinical texts. These texts, which often contain protected health information (PHI), are exposed to information extraction tools for downstream applications, risking patient identification. Existing works in de-identification rely on using large-scale annotated corpora in English, which often are not suitable in real-world multilingual settings. Pre-trained language models (LM) have shown great potential for cross-lingual transfer in low-resource settings. In this work, we empirically show the few-shot cross-lingual transfer property of LMs for named entity recognition (NER) and apply it to solve a low-resource and real-world challenge of code-mixed (Spanish-Catalan) clinical notes de-identification in the stroke domain. We annotate a gold evaluation dataset to assess few-shot setting performance where we only use a few hundred labeled examples for training. Our model improves the zero-shot F1-score from 73.7% to 91.2% on the gold evaluation set when adapting Multilingual BERT (mBERT) Devlin et al. (2019) from the MEDDOCAN Marimon et al. (2019) corpus with our few-shot cross-lingual target corpus. When generalized to an out-of-sample test set, the best model achieves a human-evaluation F1-score of 97.2%.
1 Introduction
With growing interest and innovations in data-driven digital technologies, privacy has become an important legal topic for the technology to be regulations-compliant. In Europe, the General Data Protection Regulation (GDPR) Regulation (2016) requires data owners to have a legal basis for processing personally identifiable information (PII), which also includes the explicit consent of the subjects. In cases where explicit consent is not possible, anonymization is often seen as a resorted-to solution. Clinical texts contain rich information about patients, including their gender, age, profession, residence, family, and history, that is useful for record keeping and billing purposes Johnson et al. (2016); Shickel et al. (2017).

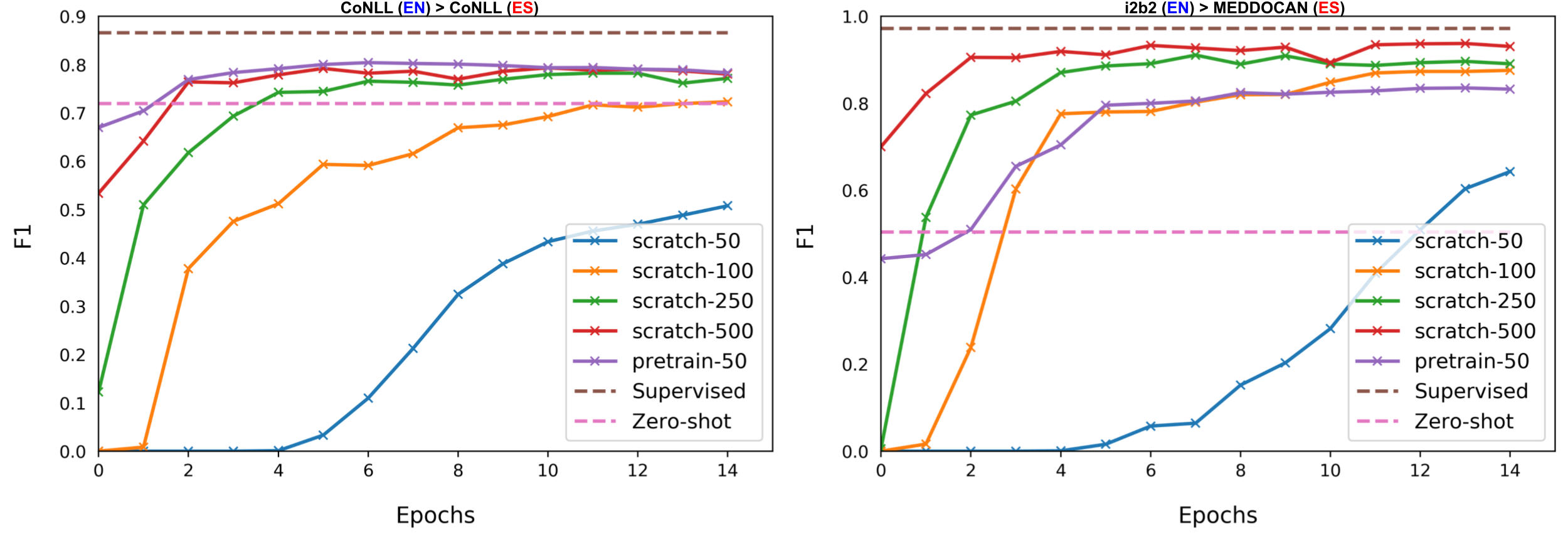
In this work, we focus on the task of removing PHI from clinical texts, also called de-identification (Fig.˜1). We address a real-world challenge where the target texts are code-mixed (Spanish-Catalan) and domain-constrained (stroke). To avoid high annotation costs, we consider a more realistic setting where we annotate a gold evaluation corpus and a few hundred examples for training. Our approach is motivated by strong performance of pre-trained LMs in few-shot cross-lingual transfer for NER with high sample efficiency (see Fig.˜2) in comparison to supervised or unsupervised approaches.
Our contributions are summarized as follows:
-
•
We empirically show the few-shot cross-lingual transfer property of multi-lingual pre-trained LM, mBERT, for NER.
-
•
We apply this property to solve a low-resource problem of code-mixed and domain-specific clinical note de-identification.
-
•
We annotate a few-shot training corpus and a gold evaluation set, minimizing annotation costs while achieving a significant performance boost without needing a large-scale labeled training corpus.
2 Related Work
GDPR-compliant anonymization requires complete and irreversible removal of any information that may lead to a subject’s data being identified (directly or indirectly) from a dataset Elliot et al. (2016). However, de-identification is limited to removing specific predefined direct identifiers; further replacement of such direct identifiers with pseudonyms is referred to as pseudonymization Alfalahi et al. (2012). Generally, de-identification can be seen as a subset of anonymization despite interchangeable usage of the terms in the literature Chevrier et al. (2019). We focus on solving the problem of de-identification in the clinical domain as a sequence labeling task, specifically named entity recognition (NER) Lample et al. (2016).
2.1 Clinical De-identification
2014 i2b2/UTHealth Stubbs and Uzuner (2015), and the 2016 CEGS N-GRID Stubbs et al. (2017) shared tasks explore the challenges of clinical de-identification on diabetic patient records and psychiatric intake records respectively. Earlier works include machine learning and rule-based approaches Meystre et al. (2010); Yogarajan et al. (2018), with Liu et al. (2017) and Dernoncourt et al. (2017) being the first to propose neural architectures. Friedrich et al. (2019) propose an adversarial approach to learn privacy-preserving text representations; Yang et al. (2019) use domain-specific embeddings trained on unlabeled corpora. While most works have mainly focused on English, some efforts have been made for Swedish Velupillai et al. (2009); Alfalahi et al. (2012) and Spanish (with a synthetic dataset at the MEDDOCAN shared task Marimon et al. (2019)).
As outlined in Lison et al. (2021), a significant challenge in clinical text de-identification is the lack of labeled data. Hartman et al. (2020) show that a small number of manually labeled PHI examples can significantly improve performance. Prior works in few-shot NER consider the problem where a model is trained on one or more source domains and tested on unseen domains with a few labeled examples per class, some of which with entity tags different from those in the source domains Yang and Katiyar (2020). Models are trained with prototypical methods, noisy supervised pre-training, or self-labeling Huang et al. (2020).
We consider a setting where the target and source domains share the same entity (PHI) tags, but with a few labeled examples in the target domain (or language). A similar setup has been employed in few-shot question answering Ram et al. (2021).
3 Problem Statement
We approach the de-identification problem as an NER task. Given an input sentence with words: , we feed it to an encoder to obtain a sequence of hidden representations
We feed into the NER classifier which is a linear classification layer with the softmax activation function to predict the PHI label of :
is the probability distribution of PHI labels for sentence and is the PHI label set. denote the set of learnable parameters and being the hidden dimension. The model is trained to minimize the per-sample negative log-likelihood:
| (1) |
For pre-trained LMs, this setting corresponds to NER fine-tuning Wu and Dredze (2019). When we jointly fine-tune on more than one NER dataset, we refer to it as multi-task learning.
Definition 1 (Few-Shot NER).
Given an entity label set , we define the task of few-shot NER as having access to labeled sentences containing each element at least once, where is a small number (e.g., in ) and is orders of magnitude larger (e.g., ).
Definition 2 (Few-Shot NER Transfer).
Given an NER dataset in a source domain (or language), we define the task of few-shot cross-domain (or cross-lingual) NER transfer as adapting a model trained on the source domain (or language) to a target domain (or language) with access to a few-shot corpus (Definition˜1).
This setting is different from prior studies in NER transfer including few-shot Huang et al. (2020), unsupervised Keung et al. (2020), and semi-supervised NER Amin and Neumann (2021).
3.1 Few-Shot Cross-Lingual Transfer
mBERT Devlin et al. (2019) has been shown to achieve strong performance for zero-shot cross-lingual transfer tasks, including NER Wu and Dredze (2019); Pires et al. (2019). Adversarial learning has been applied with limited gains Keung et al. (2019) in unsupervised approaches to improve zero-shot NER transfer, whereas feature alignments have shown better results Wang et al. (2020). Meta-learning with minimal resources Wu et al. (2020b) and word-to-word translation Wu et al. (2020a) have shown further performance gains. The current state-of-the-art approach Chen et al. (2021) combines token-level adversarial learning with self-labeled data selection and knowledge distillation.
| CoNLL (EN) CoNLL (ES) | F1 |
|---|---|
| Pires et al. (2019) | 73.59 |
| Wu and Dredze (2019) | 74.96 |
| Keung et al. (2019) | 74.30 |
| Wang et al. (2020) | 75.77 |
| Wu et al. (2020b) | 77.30 |
| Wu et al. (2020a) | 76.75 |
| Chen et al. (2021) | 79.00 |
| few-50 (or pretrain-50) | 78.30 |
To investigate the few-shot transferability of mBERT, we consider two pairs of datasets with English as the source language and Spanish as the target language: the CoNLL-2003/CoNLL-2002 Tjong Kim Sang (2002a, b) in the general domain and i2b2/MEDDOCAN Stubbs and Uzuner (2015); Marimon et al. (2019) in the clinical domain. We report results of our preliminary study in Fig.˜2. We observed that with as few as 50 random labeled training samples from the target language, we obtain substantial gains for both datasets, with near state-of-the-art on CoNLL (Table˜1). We refer to this as few-shot cross-lingual transfer property of mBERT for NER. Our study highlights that the property holds for different domains (general and clinical), where the latter focuses on the de-identification task. We leave a large-scale study on more datasets with different languages and domains as future work.
Compared to supervised (unsupervised) methods, which use complete labeled (unlabeled) target data, our few-shot approach is sample-efficient and alleviates the need of complex pipelines Wu et al. (2020b, a); Chen et al. (2021) and large-scale annotations. Furthermore, Keung et al. (2020) highlights the spurious effects of using source data as a development set and recommends using target data as a development set for model selection in NER transfer. Our findings and those in Hartman et al. (2020) motivate us to (a) propose an optimal few-shot cross-lingual transfer strategy (outlined in Fig.˜3), (b) annotate a target development set, and (c) construct an annotated few-shot target corpus for effective cross-lingual transfer learning.
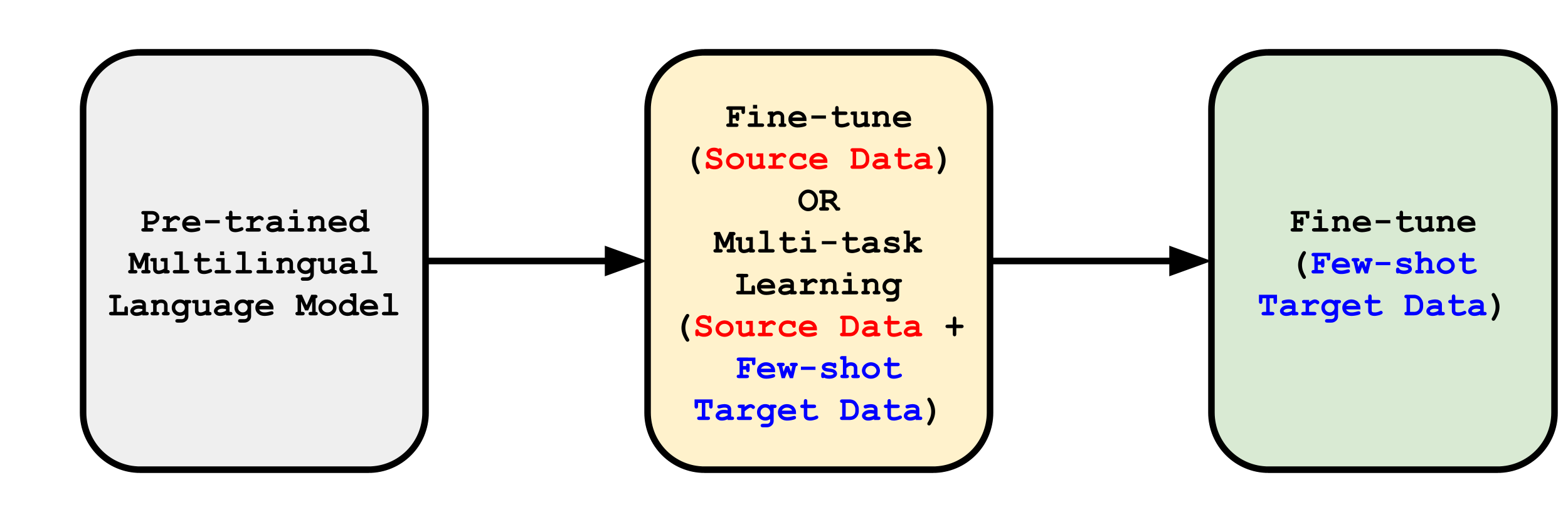
4 Data and Annotation
Our dataset consists of stroke patient records collected at Instiut Guttmann.111https://www.guttmann.com/ca/institut-universitari-guttmann-uab Table˜2 summarizes the raw data statistics and Table˜A.1 in Appendix˜A describes the topics present in the texts. We set aside 100 randomly sampled notes for out-of-sample generalizability evaluation and consider the remaining notes for our development and few-shot corpora sampling; 396 sentences are tokenized in the process.
Following the protocol in Gao et al. (2021) for constructing manually annotated distantly supervised relation extraction test sets, we train mBERT on the MEDDOCAN corpus, using coarse-grained PHI categories DATE, AGE, LOCATION, NAME, CONTACT, PROFESSION, ID with the BIO scheme Farber et al. (2008), for evaluation and few-shot training data selection. We use the trained model to make predictions on the dataset and observe that the model predicts PHI on only 50 out of the 396 sentences. A dataset of 5000 sentences ( 2% of raw sentences) is constructed from a mix of randomly sampled 2500 sentences from this 50 and 2500 from the remaining sentences. We split the dataset into two partitions of 2500 sentences for independent annotation by two annotators. The annotation is performed one sentence at a time by applying one of the 7 coarse-grained PHI labels to each token using the T2NER-annotate toolkit Amin and Neumann (2021). Each annotator’s confidence level between 1-5 is recorded for the token-level labels for each sample. To record the inter-annotator agreement, we use token-level Cohen’s kappa Cohen (1960) statistic reaching a value of 0.898. In total, the two annotators agreed on 3924 sentences, resulting in our final evaluation set. To save annotation costs for developing a few-shot target corpus, we resolved the disagreements to obtain a 384-sentence few-shot corpus for training (see Appendix˜B for annotation details).
Our source dataset (the MEDDOCAN corpus) consists of 1000 synthetically generated clinical case studies in Spanish Marimon et al. (2019). The corpus was selected manually by a practicing physician and augmented with PHI from discharge summaries and clinical records. In contrast, our target corpus focuses on the stroke domain and contains PHI from real-world records. Since the target data is code-mixed between Spanish and Catalan, with the majority (53%) being Catalan, the transfer from Spanish source data (MEDDOCAN) is cross-lingual.222Although similar, Spanish and Catalan are distinct languages. The domain of MEDDOCAN is missing an explicit mention in Marimon et al. (2019).
| Patients | Notes | ES | CA | Other |
| 1,500 | 327,775 | 42.8% | 53.0% | 4.2% |
| Transfer Strategy | Precision | Recall | F1 |
|---|---|---|---|
| Fine-tune (M) | 80.1 | 68.2 | 73.7 |
| Fine-tune (M) Fine-tune (F) | 83.5 | 94.2 | 88.6 |
| Multi-task (M + F) | 86.0 | 93.3 | 89.5 |
| Multi-task (M + F) Fine-tune (F) | 87.7 | 95.0 | 91.2 |
5 Experiments and Results
We conduct our experiments with the T2NER framework Amin and Neumann (2021).333https://github.com/suamin/T2NER For the baseline, we consider zero-shot performance on the evaluation set of the mBERT encoder fine-tuned on the MEDDOCAN training set consisting of 16,299 samples. We then fine-tune it on the few-shot target corpus as outlined in Fig.˜3. Following the multi-task learning Lin et al. (2018) approach in T2NER, we jointly fine-tune mBERT on the MEDDOCAN and few-shot target corpora. Since the few-shot corpus is much smaller, the multi-task learning helps the model transfer. It further acts as a regularization approach by sharing parameters between the datasets. To improve performance on the target data, we further fine-tune with the few-shot target corpus after the first step of fine-tuning to have improved target performance; for the model to be an expert in target Cao et al. (2020). All the models are trained for 3 epochs with a learning rate of 3e-5 and linear warm-up of 10%. For few-shot fine-tuning only, the model is trained for 25 epochs.
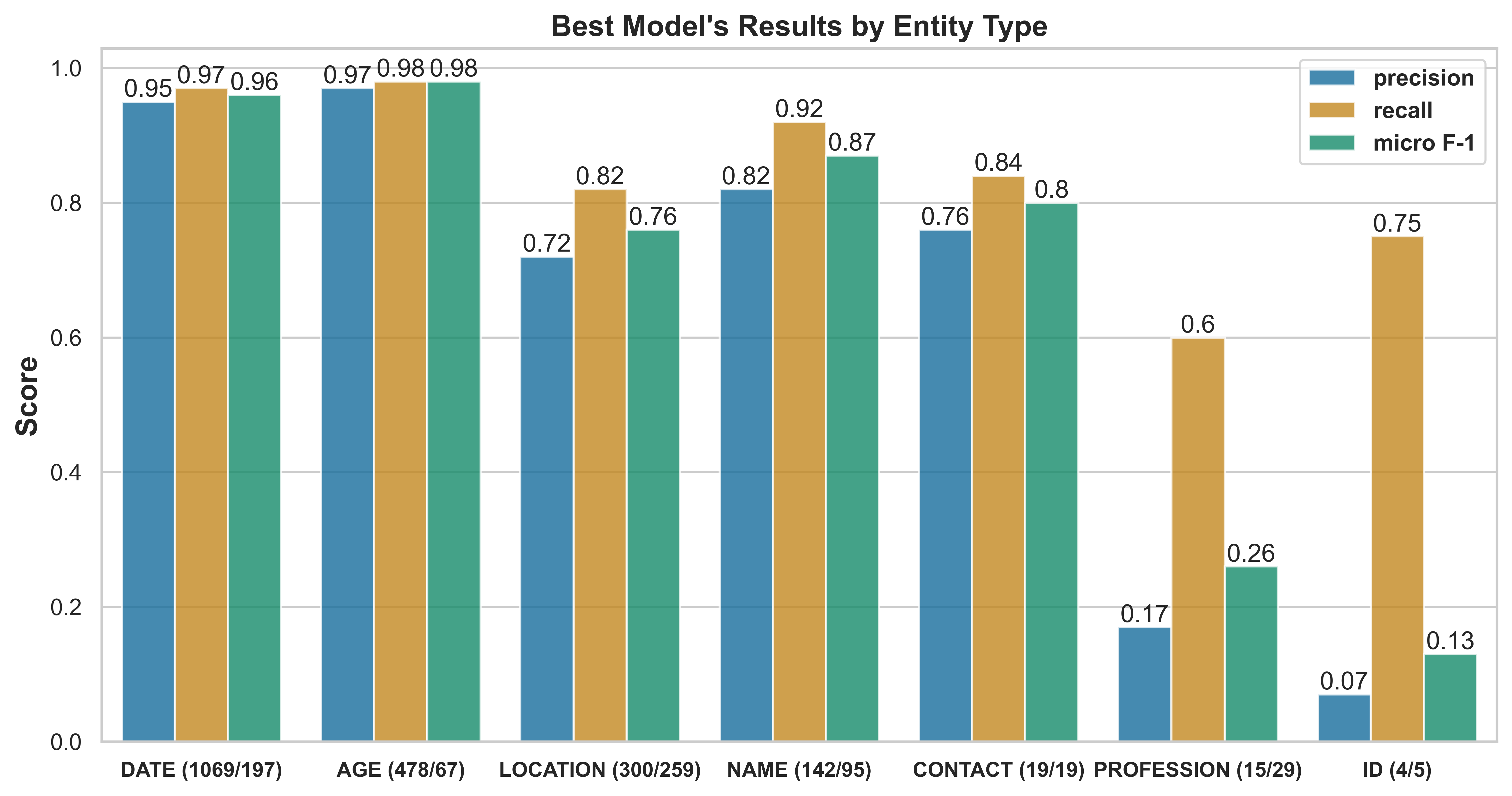
Table˜3 shows our results. Fine-tuning the baseline mBERT model with the few-shot target corpus improves the F1-score from 73.7% to 88.6%, a substantial gain of 14.9%, highlighting the effectiveness of few-shot cross-lingual transfer with mBERT. The significant increase in recall (26% points) compared to precision (3.4% points) suggests an increase in the model’s capacity to recognize domain-specific entities. Multi-task fine-tuning improves the F1-score to 89.5%; further fine-tuning on the few-shot target corpus boosts the best model’s performance to 91.2%. Fig.˜4 shows per-PHI-label scores on the development set, along with their frequency. The model performs almost perfectly on DATE and AGE, since most DATE and AGE labeled segments are similar between Spanish and Catalan as they are simple numbers (for DATE) and numbers followed by the word (for AGE; ‘edad’ in both Spanish and Catalan). There are some differences in time expressions, e.g., day of the week, as the words are distinctly dissimilar. However, structurally there is only a slight difference. Further, the model struggles with the ID class due to low sample size (5 instances in the few-shot corpus), and it is generally challenging to disambiguate between an alphanumeric string and a PHI ID, as also noted by the ID class’ high recall. Our error analysis reveals high false positives for the PROFESSION label in Catalan, e.g.: ‘Coloma de Gramenet’ (a LOCATION) and ‘Dialogant’ (being able to communicate) are both labeled as PROFESSION.
To test the model’s generalizability, we tokenize the 100 out-of-sample notes into sentences and make predictions with our best model. The resulting annotated sentences are reconstructed into patient notes, which are manually evaluated by two reviewers (one external and one annotator) for occurrences of true and false positives and negatives. The model achieves precision, recall, and F1-scores of 95.1%, 99.3%, and 97.1% respectively on the out-of-sample notes, highlighting the effectiveness of our approach.
6 Conclusion
We address the task of clinical notes de-identification in a low-resource scenario. By investigating the few-shot cross-lingual transfer property of mBERT, we propose a strategy that significantly boosts zero-shot performance while keeping the number of required annotated samples low. Our results highlight the effectiveness of the proposed strategy for the task with a potential for future applications in other low-resource scenarios.
Acknowledgments
The authors would like to thank the anonymous reviewers and Josef van Genabith for their helpful feedback. The work was partially funded by the European Union (EU) Horizon 2020 research and innovation programme through the project Precise4Q (777107), the German Federal Ministry of Education and Research (BMBF) through the project CoRA4NLP (01IW20010), the grant RTI2018-099039-J-I00 funded by MCIN/AEI/10.13039/5011000110033/ and “FEDER”, by the EU. The authors also acknowledge the cluster compute resources provided by the DFKI.
References
- Alfalahi et al. (2012) Alyaa Alfalahi, Sara Brissman, and Hercules Dalianis. 2012. Pseudonymisation of personal names and other phis in an annotated clinical swedish corpus. In Third Workshop on Building and Evaluating Resources for Biomedical Text Mining (BioTxtM 2012) Held in Conjunction with LREC, pages 49–54. Citeseer.
- Amin and Neumann (2021) Saadullah Amin and Guenter Neumann. 2021. T2NER: Transformers based transfer learning framework for named entity recognition. In Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: System Demonstrations, pages 212–220, Online. Association for Computational Linguistics.
- Cao et al. (2020) Yu Cao, Meng Fang, Baosheng Yu, and Joey Tianyi Zhou. 2020. Unsupervised domain adaptation on reading comprehension. In The Thirty-Fourth AAAI Conference on Artificial Intelligence, AAAI 2020, The Thirty-Second Innovative Applications of Artificial Intelligence Conference, IAAI 2020, The Tenth AAAI Symposium on Educational Advances in Artificial Intelligence, EAAI 2020, New York, NY, USA, February 7-12, 2020, pages 7480–7487. AAAI Press.
- Chen et al. (2021) Weile Chen, Huiqiang Jiang, Qianhui Wu, Börje Karlsson, and Yi Guan. 2021. AdvPicker: Effectively Leveraging Unlabeled Data via Adversarial Discriminator for Cross-Lingual NER. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 743–753, Online. Association for Computational Linguistics.
- Chevrier et al. (2019) Raphaël Chevrier, Vasiliki Foufi, Christophe Gaudet-Blavignac, Arnaud Robert, Christian Lovis, et al. 2019. Use and understanding of anonymization and de-identification in the biomedical literature: scoping review. Journal of medical Internet research, 21(5):e13484.
- Cohen (1960) Jacob Cohen. 1960. A coefficient of agreement for nominal scales. Educational and psychological measurement, 20(1):37–46.
- Dernoncourt et al. (2017) Franck Dernoncourt, Ji Young Lee, Ozlem Uzuner, and Peter Szolovits. 2017. De-identification of patient notes with recurrent neural networks. Journal of the American Medical Informatics Association, 24(3):596–606.
- Devlin et al. (2019) Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2019. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pages 4171–4186, Minneapolis, Minnesota. Association for Computational Linguistics.
- Elliot et al. (2016) Mark Elliot, Elaine Mackey, Kieron O’Hara, and Caroline Tudor. 2016. The anonymisation decision-making framework. ukan.
- Farber et al. (2008) Benjamin Farber, Dayne Freitag, Nizar Habash, and Owen Rambow. 2008. Improving NER in Arabic using a morphological tagger. In Proceedings of the Sixth International Conference on Language Resources and Evaluation (LREC’08), Marrakech, Morocco. European Language Resources Association (ELRA).
- Friedrich et al. (2019) Max Friedrich, Arne Köhn, Gregor Wiedemann, and Chris Biemann. 2019. Adversarial learning of privacy-preserving text representations for de-identification of medical records. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pages 5829–5839, Florence, Italy. Association for Computational Linguistics.
- Gao et al. (2021) Tianyu Gao, Xu Han, Yuzhuo Bai, Keyue Qiu, Zhiyu Xie, Yankai Lin, Zhiyuan Liu, Peng Li, Maosong Sun, and Jie Zhou. 2021. Manual evaluation matters: Reviewing test protocols of distantly supervised relation extraction. In Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021, pages 1306–1318, Online. Association for Computational Linguistics.
- Gunn et al. (2004) Patrick P Gunn, Allen M Fremont, Melissa Bottrell, Lisa R Shugarman, Jolene Galegher, and Tora Bikson. 2004. The health insurance portability and accountability act privacy rule: a practical guide for researchers. Medical care, pages 321–327.
- Hartman et al. (2020) Tzvika Hartman, Michael D Howell, Jeff Dean, Shlomo Hoory, Ronit Slyper, Itay Laish, Oren Gilon, Danny Vainstein, Greg Corrado, Katherine Chou, et al. 2020. Customization scenarios for de-identification of clinical notes. BMC medical informatics and decision making, 20(1):1–9.
- Huang et al. (2020) Jiaxin Huang, Chunyuan Li, Krishan Subudhi, Damien Jose, Shobana Balakrishnan, Weizhu Chen, Baolin Peng, Jianfeng Gao, and Jiawei Han. 2020. Few-shot named entity recognition: A comprehensive study. ArXiv preprint, abs/2012.14978.
- Johnson et al. (2016) Alistair EW Johnson, Tom J Pollard, Lu Shen, Li-wei H Lehman, Mengling Feng, Mohammad Ghassemi, Benjamin Moody, Peter Szolovits, Leo Anthony Celi, and Roger G Mark. 2016. Mimic-iii, a freely accessible critical care database. Scientific data, 3(1):1–9.
- Keung et al. (2019) Phillip Keung, Yichao Lu, and Vikas Bhardwaj. 2019. Adversarial learning with contextual embeddings for zero-resource cross-lingual classification and NER. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), pages 1355–1360, Hong Kong, China. Association for Computational Linguistics.
- Keung et al. (2020) Phillip Keung, Yichao Lu, Julian Salazar, and Vikas Bhardwaj. 2020. Don’t use English dev: On the zero-shot cross-lingual evaluation of contextual embeddings. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages 549–554, Online. Association for Computational Linguistics.
- Lample et al. (2016) Guillaume Lample, Miguel Ballesteros, Sandeep Subramanian, Kazuya Kawakami, and Chris Dyer. 2016. Neural architectures for named entity recognition. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, pages 260–270, San Diego, California. Association for Computational Linguistics.
- Lin et al. (2018) Ying Lin, Shengqi Yang, Veselin Stoyanov, and Heng Ji. 2018. A multi-lingual multi-task architecture for low-resource sequence labeling. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 799–809, Melbourne, Australia. Association for Computational Linguistics.
- Lison et al. (2021) Pierre Lison, Ildikó Pilán, David Sanchez, Montserrat Batet, and Lilja Øvrelid. 2021. Anonymisation models for text data: State of the art, challenges and future directions. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 4188–4203, Online. Association for Computational Linguistics.
- Liu et al. (2017) Zengjian Liu, Buzhou Tang, Xiaolong Wang, and Qingcai Chen. 2017. De-identification of clinical notes via recurrent neural network and conditional random field. Journal of biomedical informatics, 75:S34–S42.
- Marimon et al. (2019) Montserrat Marimon, Aitor Gonzalez-Agirre, Ander Intxaurrondo, Heidy Rodriguez, Jose Lopez Martin, Marta Villegas, and Martin Krallinger. 2019. Automatic de-identification of medical texts in spanish: the meddocan track, corpus, guidelines, methods and evaluation of results. In IberLEF@ SEPLN, pages 618–638.
- Meystre et al. (2010) Stephane M Meystre, F Jeffrey Friedlin, Brett R South, Shuying Shen, and Matthew H Samore. 2010. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC medical research methodology, 10(1):1–16.
- Pires et al. (2019) Telmo Pires, Eva Schlinger, and Dan Garrette. 2019. How multilingual is multilingual BERT? In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pages 4996–5001, Florence, Italy. Association for Computational Linguistics.
- Ram et al. (2021) Ori Ram, Yuval Kirstain, Jonathan Berant, Amir Globerson, and Omer Levy. 2021. Few-shot question answering by pretraining span selection. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 3066–3079, Online. Association for Computational Linguistics.
- Regulation (2016) Protection Regulation. 2016. Regulation (eu) 2016/679 of the european parliament and of the council. Regulation (eu), 679:2016.
- Shickel et al. (2017) Benjamin Shickel, Patrick James Tighe, Azra Bihorac, and Parisa Rashidi. 2017. Deep ehr: a survey of recent advances in deep learning techniques for electronic health record (ehr) analysis. IEEE journal of biomedical and health informatics, 22(5):1589–1604.
- Stubbs et al. (2017) Amber Stubbs, Michele Filannino, and Özlem Uzuner. 2017. De-identification of psychiatric intake records: Overview of 2016 cegs n-grid shared tasks track 1. Journal of biomedical informatics, 75:S4–S18.
- Stubbs and Uzuner (2015) Amber Stubbs and Özlem Uzuner. 2015. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/uthealth corpus. Journal of biomedical informatics, 58:S20–S29.
- Tjong Kim Sang (2002a) Erik F. Tjong Kim Sang. 2002a. Introduction to the CoNLL-2002 shared task: Language-independent named entity recognition. In COLING-02: The 6th Conference on Natural Language Learning 2002 (CoNLL-2002).
- Tjong Kim Sang (2002b) Erik F. Tjong Kim Sang. 2002b. Introduction to the CoNLL-2002 shared task: Language-independent named entity recognition. In COLING-02: The 6th Conference on Natural Language Learning 2002 (CoNLL-2002).
- Velupillai et al. (2009) Sumithra Velupillai, Hercules Dalianis, Martin Hassel, and Gunnar H Nilsson. 2009. Developing a standard for de-identifying electronic patient records written in swedish: precision, recall and f-measure in a manual and computerized annotation trial. International journal of medical informatics, 78(12):e19–e26.
- Wang et al. (2020) Zirui Wang, Jiateng Xie, Ruochen Xu, Yiming Yang, Graham Neubig, and Jaime G. Carbonell. 2020. Cross-lingual alignment vs joint training: A comparative study and A simple unified framework. In 8th International Conference on Learning Representations, ICLR 2020, Addis Ababa, Ethiopia, April 26-30, 2020. OpenReview.net.
- Wu et al. (2020a) Qianhui Wu, Zijia Lin, Börje F. Karlsson, Biqing Huang, and Jianguang Lou. 2020a. Unitrans : Unifying model transfer and data transfer for cross-lingual named entity recognition with unlabeled data. In Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, IJCAI 2020, pages 3926–3932. ijcai.org.
- Wu et al. (2020b) Qianhui Wu, Zijia Lin, Guoxin Wang, Hui Chen, Börje F Karlsson, Biqing Huang, and Chin-Yew Lin. 2020b. Enhanced meta-learning for cross-lingual named entity recognition with minimal resources. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 34, pages 9274–9281.
- Wu and Dredze (2019) Shijie Wu and Mark Dredze. 2019. Beto, bentz, becas: The surprising cross-lingual effectiveness of BERT. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), pages 833–844, Hong Kong, China. Association for Computational Linguistics.
- Yang et al. (2019) Xi Yang, Tianchen Lyu, Qian Li, Chih-Yin Lee, Jiang Bian, William R Hogan, and Yonghui Wu. 2019. A study of deep learning methods for de-identification of clinical notes in cross-institute settings. BMC Medical Informatics and Decision Making, 19(5):1–9.
- Yang and Katiyar (2020) Yi Yang and Arzoo Katiyar. 2020. Simple and effective few-shot named entity recognition with structured nearest neighbor learning. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages 6365–6375, Online. Association for Computational Linguistics.
- Yogarajan et al. (2018) Vithya Yogarajan, Michael Mayo, and Bernhard Pfahringer. 2018. A survey of automatic de-identification of longitudinal clinical narratives. ArXiv preprint, abs/1810.06765.
Appendix A Additional Dataset Details
In addition to the 7 coarse-grained PHI entities in Stubbs and Uzuner (2015), our dataset contains cross-sentence recurring entities about topics that may be of interest in the clinical domain. These topics are grouped by their potential clinical application areas and are summarized in Table˜A.1.
The label frequency distribution, as noted in Fig.˜4, is consistent with general characteristics of medical notes, which usually highlight notable events such as symptom onsets, procedures, admissions, transfers, and discharges, in addition to the date of each documentation. As a result, they tend to contain a higher frequency for the DATE PHI. In addition, the lower occurrence of the NAME PHI compared to the AGE and LOCATION entities is consistent with how healthcare providers usually communicate patient’s information.
Healthcare providers are trained to refer to patients simply by their age, gender, and the appropriate diagnosis to avoid inadvertently sharing HIPAA-sensitive information, e.g., "a 60-year-old male with ischemic stroke admitted on [DATE] from [LOCATION] (…)". The patient’s name may be used at the beginning of a medical note; however, subsequent anaphoric references are often accomplished via pronouns, omitting the NAME entity in the process. In addition, as it is applicable to Spanish medical records, nominative pronouns anaphorically referencing a patient may be omitted as they are grammatical in Spanish.
We avoid releasing our dataset due to presence of real PHI information. We will consider replacing the real PHI with synthetic ones, similar to MEDDOCAN, for a GDPR-compliant release.
Topic Areas Subcategories Diagnostics & Treatments Ischemic vs Hemorrhagic Affected areas and vessels Comorbidities Medication history Associated lifestyle factors Treatments and interventions Symptoms & Monitoring Vital signs Lab results and cultures Pain and comfort Bladder and bowel controls Long-term Care & Discharge Planning Mobility Cognitive ability Nutrition Psychosocial factors
Appendix B Annotation
B.1 Annotator Profile
Two graduate research assistants completed the annotation of the dataset. Both annotators have at a minimum CEFR444https://www.coe.int/en/ B2-C1 Spanish (Castilian) proficiency. One annotator also has clinical experience in the cardiovascular and cerebrovascular specialty, including knowledge of Spanish medical terminology. Neither annotator has formal training in Catalan; both have prior experience working with text data in the language in this domain.
B.2 Annotation Guidelines
The annotation process followed criteria for each entity as described in Stubbs and Uzuner (2015). The 7 entities: AGE, CONTACT, DATE, ID, LOCATION, NAME, and PROFESSION represent a larger granularity of the 18 HIPAA-defined PHI Stubbs and Uzuner (2015). We examined the training sets of i2b2 Stubbs and Uzuner (2015) and MEDDOCAN Marimon et al. (2019) and adapted the i2b2 annotation guidelines to create our own annotation guidelines. This step was necessary since we only focused on coarse-grained PHI types compared to fine-grained types considered in these two datasets. The adjusted guidelines utilized in this annotation process are summarized in Table˜A.5.
Description Observation Sentences annotated by annotator (A) 4400 Sentences annotated by annotator (B) 4343 Sentences annotated and revised by (A, B) 4314 Agreements 3924 Disagreements 390 Token-level Cohen’s Kappa score 0.898 Development Corpus 3924 w entity mentions 1493 w/o entity mentions 2431 Few-Shot Target Corpus 384 w entity mentions 369 w/o entity mentions 15
B.3 Annotation Procedure
Both annotators reviewed and revised their work without discussion or knowledge of the other annotator’s work.
In cross-revision, the reviewing annotator only made corrections when labeling inconsistencies were due to a lack of medical terminology comprehension.
During revision, no changes to the original annotator’s confidence level rating were made.
Confidence Level: The criteria for the confidence levels are annotator dependent as summarized in Table˜A.6 with examples.
PHI has been manually modified from the original data to preserve privacy while maintaining exemplary characteristics for each label entity type.
Skipped Sentences: Each annotator followed an independent set of criteria to exclude sentences from annotation, as demonstrated by examples in Table˜A.3.
Annotator Sample sentence Explanation A “Actualmente reside en XXXX-Xxxx Xxxxxxxx, Treballadora Social.” [Currently resides in XXXX-Xxxxx Xxxxxxxx, social worker.] The underlined words are grouped as a single word token. From the context it’s clear that ‘XXXX’ belongs to ‘LOCATION’ and ‘Xxxxx Xxxxxxxx’ are ‘NAME’ entities. B “Lmarxa. [sic]” [March or walks] Annotator does not have enough context to understand this token to annotate.
B.4 Annotator Disagreements and Resolution
An attempt was made to review the 390 sentences where our annotators disagreed to find a resolution. Main sources of disagreement are due to: (a) annotation criteria discrepancy, (b) ambiguity between related entities, and (c) annotation errors. After further revision to correct identified errors and clarify ambiguous annotation criteria, agreement was reached for 384 sentences while 6 sentences were left un-annotated due to insufficient context. Confidence levels from both annotators were left unchanged. These sentences constitute our few-shot target corpus in the pipeline explained in Fig.˜3.
Inclusion Criteria Discrepancy:
Most disagreements are related to discrepancy in the inclusion of surrounding words such as determiners, punctuation marks, and descriptive phrases. This is prevalent particularly in the LOCATION and PROFESSION entities. One annotator considered denoted sentences with these characteristics a lower confidence level of 4 compared to sentences without determiners or punctuation marks surrounding LOCATION tokens. The resolution step changed the annotations to be more consistent with the annotation guidelines described in Table˜A.5.
Ambiguity Between Related Entities:
Another source of disagreements in the LOCATION PHI stems from abbreviation usage and confusion with the NAME PHI. In instances where the syntax is ambiguous, annotators may not be able to infer correctly that certain unknown abbreviations are place names. Since it is common that places are named after people’s names and vice versa, lack of contextual information creates unresolvable ambiguity regarding the NAME and LOCATION entities. DATE and AGE also demonstrate a similar disagreement behavior. In particular, numerical and text expressions involving ‘years’ may express age or time depending on context.
Annotation Errors:
A few disagreements are due to mislabeling or erroneous omissions. There are fewer than 5 instances in the 390 disagreements. Notable errors are associated with mislabeling proper names that resemble valid named entities. For instance, some assessment tools are named after people or place names e.g. Barcelona Test and Boston (Naming) Test.
| PHI | Fine-grained Types |
|---|---|
| AGE | EDAD_SUJETO_ASISTENCIA |
| CONTACT | NUMERO_TELEFONO, NUMERO_FAX, CORREO_ELECTRONICO, URL_WEB |
| DATE | FECHAS |
| ID | ID_ASEGURAMIENTO, ID_CONTACTO_ASISTENCIAL, NUMERO_BENEF_PLAN_SALUD, |
| IDENTIF_VEHICULOS_NRSERIE_PLACAS, IDENTIF_DISPOSITIVOS_NRSERIE, | |
| IDENTIF_BIOMETRICOS, ID_SUJETO_ASISTENCIA, ID_TITULACION_PERSONAL_SANITARIO, | |
| ID_EMPLEO_PERSONAL_SANITARIO, OTRO_NUMERO_IDENTIF | |
| LOCATION | HOSPITAL, INSTITUCION, CALLE, TERRITORIO, PAIS, CENTRO_SALUD |
| NAME | NOMBRE_SUJETO_ASISTENCIA, NOMBRE_PERSONAL_SANITARIO |
| PROFESSION | PROFESION |
| OTHER | SEXO_SUJETO_ASISTENCIA, FAMILIARES_SUJETO_ASISTENCIA, |
| OTROS_SUJETO_ASISTENCIA, DIREC_PROT_INTERNET |
Appendix C MEDDOCAN Normalization
The original MEDDOCAN dataset Marimon et al. (2019) provides document level de-identification annotations, following 2014 i2b2/UTHealth Stubbs and Uzuner (2015), of 1000 clinical notes which are divided into 500, 250 and 250 for training, validation and testing respectively. It contains 29 fine-grained entity types classified into 8 coarse-grained PHI types (Table˜A.4). Compared to i2b2 (2014), MEDDOCAN has an additional OTHER category which we normalize to O in the BIO scheme, resulting in 7 coarse-grained PHI types considered in this work. We tokenize the 500 training notes resulting in 16,299 sentences. The conversion script is available in T2NER. 555https://github.com/suamin/T2NER/blob/master/utils/convert_i2b2style_xml_to_conll.py
PHI Criteria AGE Annotate only the numerical part of the expression; include both numerical and word expressions of age (e.g. 36 or thirty-six ). Include the words ‘years’, ‘months’, and ‘days’ when they express age. Include expressions that describe an age group e.g. ‘adolescent’, ‘recently born’, ‘new born’. Include punctuation associated with age, including separate tokens, e.g. in his/her 30’s. CONTACT All forms of contact information, e.g. pager, phone numbers, e-mail address. Physical or mailing address is annotated as ‘Location’ Include punctuation and symbols that occur with contact information, e.g. include all tokens in ‘(123) 456-789’. DATE Include days of the week and months. Include punctuation in all formats. Include the word ‘year’ and ‘month’ that are part of a date-time expression, e.g. ‘the year 2000’. Include prepositions that are part of a date-time expression, e.g. include the word ‘of’ in ‘5th of May’. ID Include all identification numbers such as Medical Record Number (MRN), Social Security Number (SSN), Document ID, device lot number, etc. Include any alpha-numeric expressions appearing in the beginning of the document or next to a name that’s not formatted as a date. When separated by punctuation, annotate all parts of the expression including punctuation, e.g. include all tokens in ‘12-34-5678’. Exclude the ID descriptive words and associated punctuation, e.g. exclude ‘MRN’ and ‘:’ in ‘MRN: 1234567’. LOCATION Include all place names and all parts of an address: street name, city, state, county, province, region, and country. Include punctuation in address. Include Zip/postal codes. Include organization names. Include words that specify location when they appear as part of a ‘Location’ entity, e.g. include the word ‘Center’ in ‘Social Security Center’. NAME Include only the person’s names. Include punctuation between first and last names when present Exclude titles and salutations. PROFESSION Include all professional titles, e.g. annotate ‘MD’ in the phrase ‘X works as an MD’. Exclude professional titles in name suffixes, e.g. exclude ‘MD’ in the phrase ‘Dr. X Y, MD’. Include professional and occupational descriptions, e.g. annotate ‘carpentry in the phrase ‘X works in carpentry’. Annotate the entire expression describing a profession, e.g. annotate all tokens in a phrase such as ‘worker in a cafeteria’. Exclude workplace names; annotate workplace names as ‘Location’ instead.
Level Annotator Criteria Sample Sentence Explanation 1 A Annotator is unable to assign labels due to insufficient contextual information from the given sentence. “PASe." [PASe.] or [ENTer.] The token may be an unknown acronym or an oddly typed imperative form of the verb “to enter". Insufficient context. B Annotator is unable to assign labels due to lack of comprehension. “Allitat, en DDLL.” [n/a] Annotator did not understand this sentence in Catalan sufficiently to annotate. 2 A Annotator is unsure about the assigned labels due to contextual ambiguity. ““50 años.” [50 years] Without any surrounding context, the years can be ‘AGE’ or a temporal expression; annotator thinks it’s most likely to be AGE, but does not feel confident enough to make a determination. B Annotator is unsure about the assigned labels due to lack of medical knowledge or terminology. “Urocultiu [sic] 13.01: + per A. baumanii multiR.” Urine Culture 13.01: + for MDR A. baumanii Annotator omitted this sentence due to uncertainty about the word A. baumanii, whether it could be a NAME or a non-labelled entity. 3 A Annotator is confident about the labels, but some context may be missing that could change the entity labels. “712345678)." This is likely a phone number CONTACT, but may also be an ID entity. B Annotator is confident about the sentence in general, but has some doubt due to presumed lack of specialized knowledge. “hipoTA [sic] asintomática." [Asymptomatic hypotension.] Annotator did not specify any label but was unsure whether there was a labelled entity or not. 4 A Annotator is confident about the labels, but the sentence may have some inconsistencies with the gold standard sentences. "El marido la vió y llamó a la ambulancia e ingresó en el hospital de Xxxxxxx." [The spouse saw her and called the ambulance and she was admitted to Xxxxxxx hospital.] Annotator was unsure whether to only annotate ‘Xxxxxxx’ or ‘hospital de Xxxxxxx’ or ‘el hospital de Xxxxxxx’ as LOCATION B Annotator is confident about the labels, but the sentence may have some inconsistencies with the gold standard sample sentences. “Torna d’Oftalmologia de Xxx Xxxx ( Dra. [sic]" [Returns from Xxx Xxxx Ophthalmology (Dra. ] Annotation unsure whether or not to include Ophthalmology as part of ‘LOCATION’ 5 A Annotator is confident and there’s no ambiguity regarding name entities of the labels. This could mean that the sentences have no entities to be annotated or that all the entities needing annotations are consistent with the gold standard sample sentences. “Cito a control el próximo 25.12.20 y doy pautas a la esposa." [I make a follow-up appointment for the upcoming date 25.12.20 and I give the prescription to the wife.] It’s clear that ‘25.12.20’ is a DATE PHI. B It is clear to the annotator that the sentence has no entities to be annotated or that the entities are consistent with gold standard annotation. This could be either apparent at first glance or because the sentence has been seen several times before, which increases the annotator’s confidence regarding the assigned label(s). “Cito a control el próximo 25.12.20 y doy pautas a la esposa." [I make a follow-up appointment for the upcoming date 25.12.20 and I give the prescription to the wife.] It’s clear that ‘25.12.20’ is a DATE PHI.