Improving Precancerous Case Characterization via Transformer-based Ensemble Learning
Abstract
The application of natural language processing (NLP) to cancer pathology reports has been focused on detecting cancer cases, largely ignoring precancerous cases. Improving the characterization of precancerous adenomas assists in developing diagnostic tests for early cancer detection and prevention, especially for colorectal cancer (CRC). Here we developed transformer-based deep neural network NLP models to perform the CRC phenotyping, with the goal of extracting precancerous lesion attributes and distinguishing cancer and precancerous cases. We achieved 0.914 macro-F1 scores for classifying patients into negative, non-advanced adenoma, advanced adenoma and CRC. We further improved the performance to 0.923 using an ensemble of classifiers for cancer status classification and lesion size named entity recognition (NER). Our results demonstrated the potential of using NLP to leverage real-world health record data to facilitate the development of diagnostic tests for early cancer prevention.
1 Introduction
Cancer has been the second leading cause of death with more than 1,900k new cases and 600k cancer deaths in the United States in 2022 (Siegel et al., 2019). Among those, colorectal cancer (CRC) is the third most common cancer and the third leading cause of cancer death (Siegel et al., 2019). Detecting CRC at its early stage can dramatically improve clinical outcomes. The 5-year survival rate is 90% when colorectal cancer is identified at the localized stage compared to 73% and 17% survival rates at the regional or distant stage, respectively111https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/survival-rates.html.
CRC progresses from asymptomatic non-advanced adenoma (NAA) to advanced adenoma (AA) and then to invasive carcinoma (Junca et al., 2020). AAs are adenomas characterized by villous or tubulovillous histology, adenomas or sessile serrated lesions mm, or high-grade dysplasia Junca et al. (2020); Shaukat et al. (2021). AA indicates an intermediate or high risk for CRC Lieberman et al. (2012) and requires CRC screening every three years Lieberman et al. (2012). Recent economic studies suggest a test with increasing adenoma sensitivity in a blood-based CRC screening test can reduce CRC incidence and reduce mortality Putcha et al. (2022a).
There is great interest in developing noninvasive diagnostic tests with high sensitivity and specificity for advanced adenoma and CRC screening Imperiale et al. (2014); Putcha et al. (2022a). This development process requires biomarker discovery and clinical validation based on samples collected from large numbers of individuals whose colorectal cancer statuses are confirmed by colonoscopy Putcha et al. (2022b). Correctly classifying the colorectal cancer statuses, namely negative (NEG), NAA, AA and CRC, requires expertise in distilling and interpreting tumor stage and histology information and size of precancerous adenoma from colonoscopy and pathology reports. Such nuanced annotations are typically not documented and collected in structured sections of electronic health records or standardized via International Classification of Diseases (ICD) codes Raju et al. (2015, 2013). Therefore, extracting this information from colonoscopy and pathology reports and generating reliable CRC status classification has heavily relied on manual review by trained gastrointestinal pathologists. Such review is time-consuming, costly and difficult to scale.
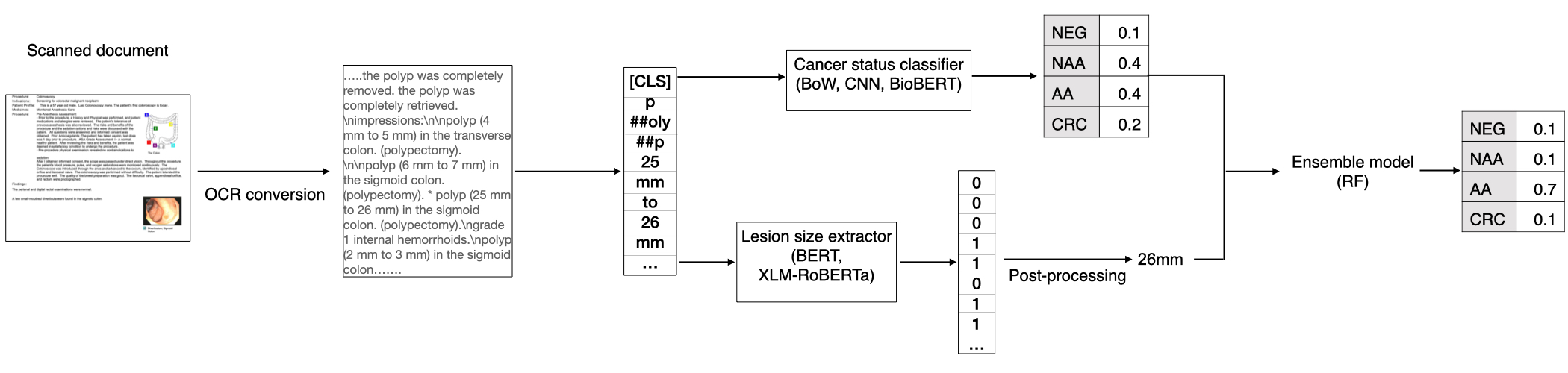
To reduce the burden of manually annotating thousands to hundreds of thousands of pathology reports, and to facilitate the development of noninvasive diagnostic tools for colorectal cancer prevention, we investigated classical and advanced natural language processing (NLP) methods to automatically extract precancerous lesion information and determine CRC status (Figure 1). We developed transformer models to extract both categorical and numerical attributes from colonoscopy and pathology reports. Compared to Bag-of-Word (BoW) and convolutional neural network (CNN) models (see Data and methods in section 3), we achieved the best performance by fine-tuning the BioBERT model. Since lesion size is an important factor to distinguish between the AA and NAA classes (Appendix A.1, Winawer and Zauber (2002)), we developed an entity recognition model for lesion size extraction and improved its performance through transfer learning from a non-biomedical domain. We further improved the cancer status classification model performance by explicitly adding extracted lesion size through an ensemble model. Our study also addressed two challenges for NLP practice that are specific to the biomedical industry setting: annotation at the sentence level for numerical variable extraction is limited; and most clinical trial studies that enroll patients from various sites still receive health records in the scanned PDF format Raju et al. (2015), creating challenges for precisely locating the diagnosis section in health records. Our research demonstrated that, along with domain knowledge-informed feature learning, fine-tuned advanced deep learning methods are able to achieve high accuracy in highly complex and nuanced disease phenotyping tasks, even with only several thousands of documents for model training.
2 Related Work
NLP methods have been applied to pathology reports to extract categorical attributes associated with cancer diagnosis. Yala et al. (2017) used machine learning methods with Bag-of-Words features to classify patients into breast cancer carcinoma and atypia categories. Adding clinical concepts from the Unified Medical Language System (UMLS) was shown to improve classification performance Li and Martinez (2010); Martinez and Li (2011). Preston et al. (2022) used embedding vectors that are pre-trained in BERT-based Devlin et al. (2018) models for tumor site, histology and TNM staging (T: tumor size/location; N: lymph node status; M: metastasis) classification from longitudinal reports and developed classifiers to detect cancer cases. Park et al. (2021) extracted cancer histology, site and surgical procedure from colon, lung and kidney cancer data. They demonstrated good performance by leveraging transfer learning across cancer types and few-shot learning by accounting for semantic similarity. Other deep learning approaches such as hierarchical attention neural networks Gao et al. (2018, 2019), multitask learning Alawad et al. (2020), and graph convolutional networks Wu et al. (2020) have been employed to extract cancer characteristics such as primary site and histological grade. However, these studies primarily focused on extracting categorical cancer characteristics that are routinely collected in cancer registries Klein and Havener (2011) and largely ignored numerical and precancerous attributes, which are critical for developing early cancer detection technology.
The extraction of numerical cancer attributes is challenging because the semantic context for numerical variables is mostly at the sentence level instead of document/patient level Li and Martinez (2010); Odisho et al. (2020). This creates a discrepancy between training objectives (sentence level) and output evaluation (patient level). To overcome this limitation, Li and Martinez (2010) first identified the sentences that contain the numerical values and extracted them through regular expression matching. AAlAbdulsalam et al. (2018) treated TNM staging extraction as a sequence labeling task with pattern matching and conditional random field techniques. Odisho et al. (2020) encoded tokens and their context words as bag-of-n-gram features and classified the token sequence for TNM staging and tumor volume extraction. In our work, we treated numerical lesion size extraction as a named entity recognition (NER) task and addressed the challenge of limited annotated sample size by transfer learning from models pre-trained in a non-biomedical domain.
Previous attempts to employ NLP methods to parse colonoscopy reports and linked pathology reports have aimed to characterize adenomas due to their importance in estimating colorectal cancer risk Lee et al. (2019); Raju et al. (2013, 2015); Imler et al. (2013). The limitations of current studies are two fold: first, most studies still rely on rule-based systems such as Linguamatics Lee et al. (2019) and cTAKEs Imler et al. (2013); Savova et al. (2010) for extracting adenomas through pattern-matching and dictionary look-up. Deriving rules can be time-consuming, require extensive domain knowledge, and likely results in overfitting to the development dataset and limited portability; second, NLP studies for colorectal cancer do not perform end-to-end CRC phenotyping to classify patients into NEG, NAA, AA and CRC, which are of great interest to characterize CRC risk and prioritize patients for cancer screening and early cancer detection. Here we performed end-to-end precancerous and cancer status characterization with deep learning methods which promise to be more generalizable and efficient than rule-based approaches.
3 Data and methods
3.1 Dataset
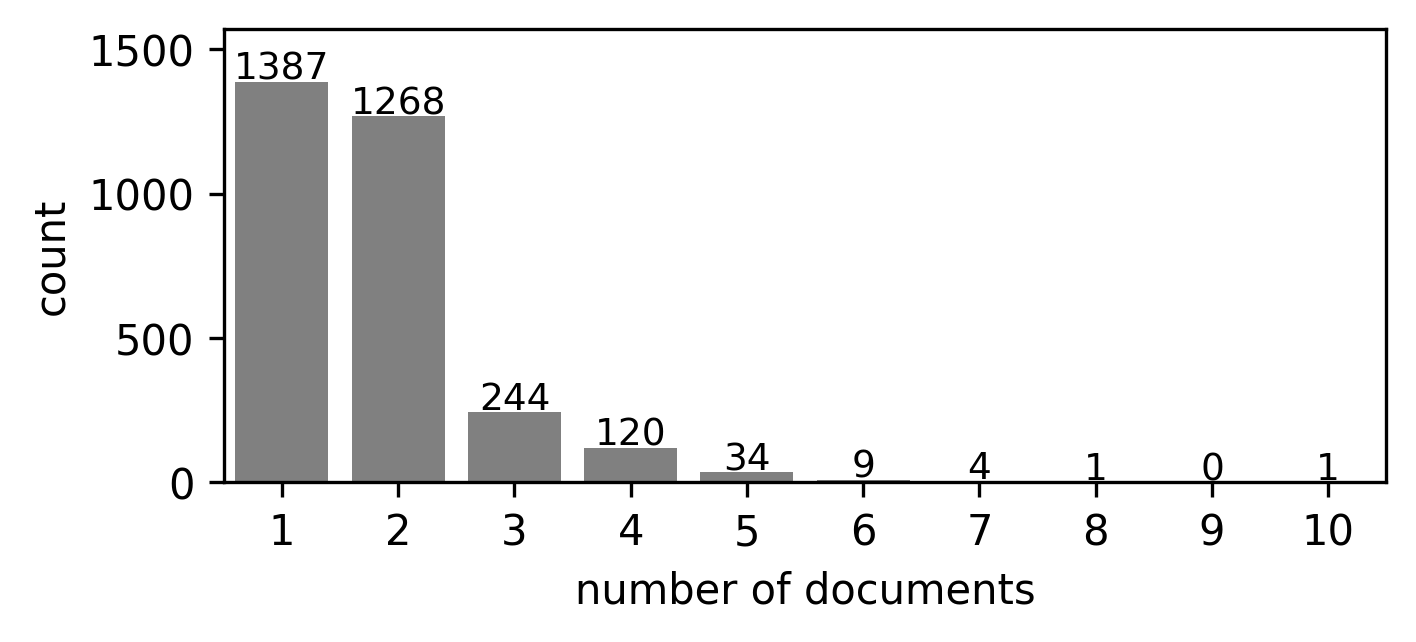
In this study, we used health records from 3,068 patients collected as part of two studies from 68 collection sites. In some cases, multiple types of health records are associated with one patient, including colonoscopy, pathology, surgical pathology, and radiology reports. In total, there are 5,405 documents for all patients. Appendix Figure A1 shows the distribution of document numbers for each patient.
We split patients into train and test sets stratified by cancer status. To assess the generalizability of NLP models when applied to pathology reports collected in unseen sites, we used samples from independent collection sites for the train and test sets. There are 2,149 samples in the training set from 54 sites and 919 samples in the test set from 14 independent sites. Appendix Table A1 shows the sample count for each cancer status in the train and test sets.
3.2 Document and sentence level annotations
A certified pathologist reviewed and assigned patient-level labels for colorectal cancer status and lesion size. The detailed annotation criteria are described in Appendix A.1. For lesion size annotation, the pathologist first identified the index lesion, which is the most clinically significant lesion according to cancer status and lesion type. Then the size of the index lesion was used as the patient-level lesion size annotation. We used zero as the lesion size for healthy samples with no identified lesions.
We generated sentence-level annotation for lesion size. We treated the lesion size named entity annotation as a binary label with tokens within the lesion size entity as 1 and tokens outside the lesion size entity as 0. We did not distinguish the start or end token of the named entity. We randomly selected 499 documents from 225 patients from the training set and 331 documents from 114 patients from the test set for NER model training and evaluation, respectively.
3.3 Data preprocessing
Since the reports are in scanned PDF format, we first digitized the reports with optical character recognition provided by the Google Vision API222https://cloud.google.com/vision/docs/ocr. The OCR algorithm outputs the recognized text and the coordinates of the bounding box for each text block.
We used fuzzy matching for words: "diagnosis," "finding," "impression," "diagnoses," "findings,", "impressions," and "polyp" with text in each bounding box. This allowed us to identify sections important for diagnosis and ignore irrelevant sections to increase the signal-to-noise ratio. To allow for some error in bounding box identification, we retained texts within 10 bounding boxes and at most 100 words after the first matched bounding box. These keywords for fuzzy matching and window size were determined by an iterative manual inspection of reports in the training set.
For CNN and BoW, we concatenated documents corresponding to one patient into one text segment and padded the concatenated text segment to the max length. For the BERT models with a sequence length limit of 512 tokens, we split the text into segments with 10 overlapping tokens.
3.4 Cancer status classification model
We built and tested three models, namely Bag-of-Words (BoW) Zhong et al. (2018), CNN Kim (2019) and BioBERT Lee et al. (2020), for cancer status classification. We split the 2,149 documents reserved for training into training and validation sets in a 9:1 ratio and selected the best model based on the validation macro-F1 score [See 3.7]. For BoW, tf-idf representation Zhong et al. (2018), a term-frequency based featurization, was derived as input features for SVM models with linear, polynomial or radial basis function (RBF) kernels. We used unigram features and removed terms that appear in less than 10 documents.
For the CNN model, instead of doing hand-crafted feature engineering, 1D convolution kernels were learned to extract localized text patterns from pathology reports. The convolutional layer is followed by a max-pooling layer and a fully connected layer to classify colorectal cancer status.
For the BERT model, BioBERT Lee et al. (2020), a pre-trained biomedical language representation, was employed and fine-tuned as follows to encode the pathology reports for cancer status classification. BioBERT was pre-trained based on BERT initiated weights with biomedical domain corpora (PubMed abstracts and PMC full-text articles) and has increased performance in biomedical text mining tasks including NER, relation extraction and question-answering. We added a fully connected layer after the [CLS] embedding vector for multiclass classification Devlin et al. (2018). Because one patient can be associated with multiple documents or text segments but the cancer status label is annotated for each patient, this creates a multiple instance learning problem. We treated each patient as a bag and each text segment as an instance within the bag. We used max-pooling to get the largest softmax probability for each class across multiple text segments and renormalize with the softmax function to calculate cross-entropy loss per patient.
3.5 Lesion size extraction model
We treated the lesion size extraction as a NER task and compared two approaches. For direct fine-tuning, we used a pretrained BERT-base-uncased333https://huggingface.co/bert-base-uncased model and classified token embedding into binary labels where the positive label indicates the target named entity. We also fine-tuned a XLM_RoBERTa_base444https://huggingface.co/xlm-roberta-base model.
Observing the similarity between lesion size vs. one of the annotated named entities (QUANTITY: Measurements, as of weight or distance) from the OntoNotes5 corpus Weischedel et al. (2011), we used an XLM_RoBERTa555https://huggingface.co/asahi417/tner-xlm-roberta-base-uncased-ontonotes5 model that was previously fine-tuned on the OntoNotes5 dataset for NER of QUANTITY. We then continued to fine-tune this model on the cancer pathology dataset for lesion size extraction to explore the benefit of transfer learning. Both direct fine-tuning and transfer learning models were trained and validated based on a 7:1 split of the sentence-level annotated documents. We selected the model with the best validation F1 score and evaluated its performance on the holdout test set. The hyperpameters can be found in Appendix Table A3.
3.6 Ensemble model
We built an ensemble model with BioBERT predicted probability for each class and binarized lesion size feature (lesion size mm or not). For the model training, we used the binarized ground-truth lesion size. For model inference, we used the binarized NER-extracted lesion size. We trained a random forest model with 10 trees and max_depth=10 as the ensemble model on the training set and tested its performance on the validation and test sets for the cancer status classification task.
3.7 Metrics
For cancer status classification evaluation, we computed the precision, recall and F1 for each cancer status. We used the macro-F1= for the overall model performance metric for multi-class classification ( classes).
For NER evaluation, we count consecutive positive labeled tokens as one named entity. We identified named entities derived from ground-truth labels (total ground-truth positive, TGP) and predicted labels (total predicted positive, TPP). We counted an exact match of starting and ending index of the ground-truth and predicted entities as true positive (TP). We calculated the precision as , recall as and F1 score for the named entity recognition task.
| Model | NEG F1 (n=515) | NAA F1 (n=183) | AA F1 (n=146) | CRC F1 (n=75) | Macro-F1 |
|---|---|---|---|---|---|
| BoW | 0.941 | 0.706 | 0.758 | 0.952 | 0.839 |
| CNN | 0.971 | 0.842 | 0.773 | 0.947 | 0.883 |
| BioBERT | 0.972 | 0.888 | 0.833 | 0.962 | 0.914 |
| BioBERT+Lesion Size | 0.965 | 0.894 | 0.854 | 0.980 | 0.923 |
4 Results
4.1 Cancer status classification model performance
We treated the cancer status (CRC, AA, NAA and NEG) extraction as a document classification problem and trained BoW, CNN and BioBERT models. All models achieved over 0.8 macro-F1 scores, with the BioBERT model outperforming BoW and CNN (Table 1). In particular, all models including BoW classified CRC and NEG with high accuracy (> 0.9 F1 score) and AA and NAA with lower accuracy (< 0.8 F1 score). This suggests that unigram features are sensitive for classifying cancer and healthy patients from pathology reports but less sensitive for differentiating precancerous patients.
The CNN model (NAA F1=0.842, AA F1=0.773) improved NAA and AA performance compared to BoW (NAA F1=0.706, AA F1=0.758). This suggests that the larger kernels used in CNN improve the capture of semantics for precancerous classes compared with unigram features. The BioBERT model further improved AA and NAA performance (NAA F1=0.888, AA F1=0.833). As the model complexity and its ability to capture long-range interaction increases, the model performed better. Although the number of training samples was limited, the more complex models appear to be more generalizable.
| Predicted Label | ||||||
|---|---|---|---|---|---|---|
| NEG | NAA | AA | CRC | Total | ||
| True Label | NEG | 1195 | 8 | 15 | 0 | 1218 |
| NAA | 7 | 456 | 17 | 1 | 481 | |
| AA | 5 | 13 | 257 | 2 | 277 | |
| CRC | 1 | 0 | 0 | 172 | 173 | |
4.2 Lesion size extraction model performance
We performed an error analysis on the training and validation data to identify the source of incorrect predictions (Table 2, Appendix Table A2) for the BioBERT model. We found misclassifications were usually confusion between AA and NAA. 68.0% (17/25) of incorrect predictions for NAAs were classified as AA, and 65.0% (13/20) of incorrect predictions for AAs were classified as NAAs. Since lesion size is an important factor to distinguish between the AA and NAA classes (Appendix A.1), we proposed to explicitly add lesion size as an additional feature to improve the BERT-based cancer status classification model.
The direct fine-tuning of the BERT model for lesion size NER had low performance, potentially due to the small sample size for sentence-level annotation (test F1 score=0.202, precision=0.159 and recall=0.273, Table 3). We then evaluated the transfer learning approach, using an XLM_RoBERTa model that had been fine-tuned on the OntoNotes dataset for QUANTITY extraction. Directly applying this model to the cancer pathology dataset to extract lesion size led to an increased F1 (0.508) score, with high recall (0.703) and low precision (0.398). We next continued to train this model on the cancer pathology dataset to perform lesion size extraction. Interestingly, additional fine-tuning substantially improved the performance (F1=0.757, precision=0.761 and recall=0.753), especially the precision. This suggests that transfer learning using models fine-tuned on tasks outside the biomedical domain can substantially improve domain-specific NLP performance, even with a relatively small sample size.
| Model | Train | Val | Test | ||||||
|---|---|---|---|---|---|---|---|---|---|
| F1 | precision | recall | F1 | precision | recall | F1 | precision | recall | |
| FT_BERT | 0.316 | 0.240 | 0.450 | 0.174 | 0.142 | 0.225 | 0.202 | 0.159 | 0.273 |
| FT_XLM_RoBERTa | 0.471 | 0.372 | 0.644 | 0.259 | 0.197 | 0278 | 0.243 | 0.186 | 0.351 |
| OntoNotes_XLM_RoBERTa | 0.395 | 0.275 | 0.702 | 0.360 | 0.243 | 0.695 | 0.508 | 0.398 | 0.703 |
| TL_OntoNotes_XLM_RoBERTa | 0.933 | 0.911 | 0.956 | 0.874 | 0.856 | 0.893 | 0.757 | 0.761 | 0.753 |
4.3 Ensemble model improves cancer status classification
We then assessed the effect of explicitly adding lesion size as an additional feature to classify cancer status. The ensemble model which combined BioBERT predicted probabilities and binarized lesion size (mm or not) improved NAA performance from 0.888 to 0.894 and AA performance from 0.833 to 0.854 while maintaining the already high performance for the NEG and CRC classes (Table 1). The macro-F1 was 0.923 for the ensemble model compared to 0.914 for the BioBERT model alone. This suggests that explicitly adding features that are informed by domain knowledge can improve classification performance compared to fine-tuned transformer models alone. This approach may be particularly beneficial for applications in which training data are limited.
4.4 Integrated gradient analysis
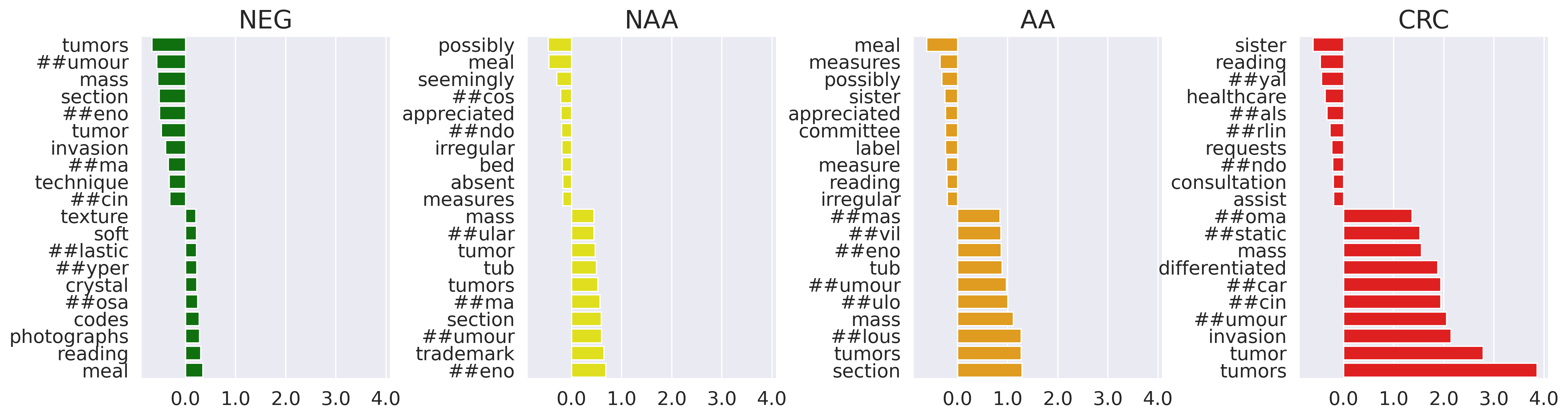
To investigate which features are most important for BioBERT model performance, we performed integrated gradient analysis, which computes attribution scores that measure feature importance with respect to the classification prediction Sundararajan et al. (2017). We calculated attribution with respect to the input embedding vector. We performed integrated gradient analysis for a random subset of 534 NEGs, 197 NAAs, 168 AAs and 80 CRCs and calculated the averaged feature attributions across documents for each class (Figure 2). High-scoring tokens related to CRC classification included “tumor,” “invasion,” and “carcinoma.” High-scoring tokens related to AA and NAA classification included “tub”, “##umour”, and “##eno.” This model interpretability analysis helped to confirm that our NLP model is able to leverage key terms that match domain knowledge.
5 Conclusion
Determining cancer status and characterizing precancerous lesions are critical and time-consuming steps for the development and evaluation of diagnostic tests for colorectal cancer screening. Here we achieved a 0.914 macro-F1 score for cancer status classification with transformer models fine-tuned using BioBERT. Informed by the domain knowledge and error analyses, we identified lesion size as a critical factor for differentiating between AAs and NAAs, but one that was not efficiently captured in BioBERT context-dependent embeddings. Using an ensemble model combining a fine-tuned BioBERT model and a lesion size named entity recognition model, we further improved classification performance to a macro-F1 score of 0.923. The lesion size extraction model was developed through transfer learning, using a transformer model trained in a non-biomedical domain. We showed that directly fine-tuning of transformer models was inadequate for domain-specific NLP tasks, and that precise feature engineering and use of ensemble models was needed to improve classification performance. Overall, we provided an accurate algorithm for characterizing precancerous cases that may help to improve early colorectal cancer detection and prevention, and a model training framework that leverages advanced NLP techniques to address complex disease phenotyping tasks in biomedical domain.
6 Limitations
One limitation of this work is that we could not fully evaluate how use of scanned reports and OCR affects performance as compared to use of electronic reports, due to a lack of dataset with paired scanned and electronic formats. The scanned format makes the selection of relevant sections from the colonoscopy and pathology reports challenging. We used fuzzy matching of selected keywords to identify sections that are likely important for diagnosis, but this process might introduce bias. Additionally, the digitization process by OCR results in errors in keywords and numerical values. For example, we observed “tubulovillous” was misrecognized as “tubulovillaus” and "0.2cm" is misrecognized as "0:2cm". This could affect the performance in NER and final cancer status classification. Future work could include evaluating other OCR tools besides Google Vision.
Another limitation is the lack of validation studies using an external dataset. It is known that health records vary substantially in both formats and content. Studies have been done to transform pathology reports to use standardized terminologies and diagnoses Kim et al. (2020); Ryu et al. (2020). Even though the dataset used in this study is collected from 68 collection sites across the US, the sample size is still relatively small and may not fully capture the variabilities of real-world data.
7 Acknowledgments
This work was supported by Freenome. We thank Paul Tittel and Michael Widrich for helpful discussion; Chuanbo Xu for assistance in acquiring the clinical documents; Amit Pasupathy for helping to collect the reports; and Anooj Patel and David Liu for support with the computational and machine learning infrastructure.
References
- AAlAbdulsalam et al. (2018) Abdulrahman K AAlAbdulsalam, Jennifer H Garvin, Andrew Redd, Marjorie E Carter, Carol Sweeny, and Stephane M Meystre. 2018. Automated extraction and classification of cancer stage mentions fromunstructured text fields in a central cancer registry. AMIA Summits on Translational Science Proceedings, 2018:16.
- Alawad et al. (2020) Mohammed Alawad, Shang Gao, John X Qiu, Hong Jun Yoon, J Blair Christian, Lynne Penberthy, Brent Mumphrey, Xiao-Cheng Wu, Linda Coyle, and Georgia Tourassi. 2020. Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks. Journal of the American Medical Informatics Association, 27(1):89–98.
- Devlin et al. (2018) Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2018. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
- Gao et al. (2019) Shang Gao, John X Qiu, Mohammed Alawad, Jacob D Hinkle, Noah Schaefferkoetter, Hong-Jun Yoon, Blair Christian, Paul A Fearn, Lynne Penberthy, Xiao-Cheng Wu, et al. 2019. Classifying cancer pathology reports with hierarchical self-attention networks. Artificial intelligence in medicine, 101:101726.
- Gao et al. (2018) Shang Gao, Michael T Young, John X Qiu, Hong-Jun Yoon, James B Christian, Paul A Fearn, Georgia D Tourassi, and Arvind Ramanthan. 2018. Hierarchical attention networks for information extraction from cancer pathology reports. Journal of the American Medical Informatics Association, 25(3):321–330.
- Imler et al. (2013) Timothy D Imler, Justin Morea, Charles Kahi, and Thomas F Imperiale. 2013. Natural language processing accurately categorizes findings from colonoscopy and pathology reports. Clinical Gastroenterology and Hepatology, 11(6):689–694.
- Imperiale et al. (2014) Thomas F Imperiale, David F Ransohoff, Steven H Itzkowitz, Theodore R Levin, Philip Lavin, Graham P Lidgard, David A Ahlquist, and Barry M Berger. 2014. Multitarget stool dna testing for colorectal-cancer screening. New England Journal of Medicine, 370(14):1287–1297.
- Junca et al. (2020) Audelaure Junca, Gaëlle Tachon, Camille Evrard, Claire Villalva, Eric Frouin, Lucie Karayan-Tapon, and David Tougeron. 2020. Detection of colorectal cancer and advanced adenoma by liquid biopsy (decalib study): The ddpcr challenge. Cancers, 12(6):1482.
- Kim et al. (2020) Baek-hui Kim, Joon Mee Kim, Gyeong Hoon Kang, Hee Jin Chang, Dong Wook Kang, Jung Ho Kim, Jeong Mo Bae, An Na Seo, Ho Sung Park, Yun Kyung Kang, et al. 2020. Standardized pathology report for colorectal cancer. Journal of pathology and translational medicine, 54(1):1–19.
- Kim (2019) Y Kim. 2019. Convolutional neural networks for sentence classification. arxiv 2014. arXiv preprint arXiv:1408.5882.
- Klein and Havener (2011) W Ted Klein and L Havener. 2011. Standards for cancer registries volume v: Pathology laboratory electronic reporting.
- Lee et al. (2019) Jeffrey K Lee, Christopher D Jensen, Theodore R Levin, Ann G Zauber, Chyke A Doubeni, Wei K Zhao, and Douglas A Corley. 2019. Accurate identification of colonoscopy quality and polyp findings using natural language processing. Journal of clinical gastroenterology, 53(1):e25.
- Lee et al. (2020) Jinhyuk Lee, Wonjin Yoon, Sungdong Kim, Donghyeon Kim, Sunkyu Kim, Chan Ho So, and Jaewoo Kang. 2020. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4):1234–1240.
- Li et al. (2017) Lisha Li, Kevin Jamieson, Giulia DeSalvo, Afshin Rostamizadeh, and Ameet Talwalkar. 2017. Hyperband: A novel bandit-based approach to hyperparameter optimization. The Journal of Machine Learning Research, 18(1):6765–6816.
- Li and Martinez (2010) Yue Li and David Martinez. 2010. Information extraction of multiple categories from pathology reports. In Proceedings of the Australasian Language Technology Association Workshop 2010, pages 41–48.
- Lieberman et al. (2012) David A Lieberman, Douglas K Rex, Sidney J Winawer, Francis M Giardiello, David A Johnson, and Theodore R Levin. 2012. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the us multi-society task force on colorectal cancer. Gastroenterology, 143(3):844–857.
- Martinez and Li (2011) David Martinez and Yue Li. 2011. Information extraction from pathology reports in a hospital setting. In Proceedings of the 20th ACM international conference on Information and knowledge management, pages 1877–1882.
- Odisho et al. (2020) Anobel Y Odisho, Briton Park, Nicholas Altieri, John DeNero, Matthew R Cooperberg, Peter R Carroll, and Bin Yu. 2020. Natural language processing systems for pathology parsing in limited data environments with uncertainty estimation. JAMIA open, 3(3):431–438.
- Park et al. (2021) Briton Park, Nicholas Altieri, John DeNero, Anobel Y Odisho, and Bin Yu. 2021. Improving natural language information extraction from cancer pathology reports using transfer learning and zero-shot string similarity. JAMIA open, 4(3):ooab085.
- Preston et al. (2022) Sam Preston, Mu Wei, Rajesh Rao, Robert Tinn, Naoto Usuyama, Michael Lucas, Roshanthi Weerasinghe, Soohee Lee, Brian Piening, Paul Tittel, et al. 2022. Towards structuring real-world data at scale: Deep learning for extracting key oncology information from clinical text with patient-level supervision. arXiv preprint arXiv:2203.10442.
- Putcha et al. (2022a) Girish Putcha, Lauren N Carroll, Tarun Chandra, and Andrew Piscitello. 2022a. Interception versus prevention in cancer screening in a medicare population: Results from the crc-maps model.
- Putcha et al. (2022b) Girish Putcha, Chuanbo Xu, MMA Shaukat, and Theodore R Levin. 2022b. Prevention of colorectal cancer through multiomics blood testing: The preempt crc study. J. Clin. Oncol, 40.
- Raju et al. (2015) Gottumukkala S Raju, Phillip J Lum, Rebecca S Slack, Selvi Thirumurthi, Patrick M Lynch, Ethan Miller, Brian R Weston, Marta L Davila, Manoop S Bhutani, Mehnaz A Shafi, et al. 2015. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointestinal endoscopy, 82(3):512–519.
- Raju et al. (2013) Gottumukkala S Raju, Vikram Vadyala, Rebecca Slack, Somashekar G Krishna, William A Ross, Patrick M Lynch, Robert S Bresalier, Ernest Hawk, and John R Stroehlein. 2013. Adenoma detection in patients undergoing a comprehensive colonoscopy screening. Cancer medicine, 2(3):391–402.
- Ryu et al. (2020) Borim Ryu, Eunsil Yoon, Seok Kim, Sejoon Lee, Hyunyoung Baek, Soyoung Yi, Hee Young Na, Ji-Won Kim, Rong-Min Baek, Hee Hwang, et al. 2020. Transformation of pathology reports into the common data model with oncology module: use case for colon cancer. Journal of medical Internet research, 22(12):e18526.
- Savova et al. (2010) Guergana K Savova, James J Masanz, Philip V Ogren, Jiaping Zheng, Sunghwan Sohn, Karin C Kipper-Schuler, and Christopher G Chute. 2010. Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications. Journal of the American Medical Informatics Association, 17(5):507–513.
- Shahriari et al. (2015) Bobak Shahriari, Kevin Swersky, Ziyu Wang, Ryan P Adams, and Nando De Freitas. 2015. Taking the human out of the loop: A review of bayesian optimization. Proceedings of the IEEE, 104(1):148–175.
- Shaukat et al. (2021) Aasma Shaukat, Charles J Kahi, Carol A Burke, Linda Rabeneck, Bryan G Sauer, and Douglas K Rex. 2021. Acg clinical guidelines: colorectal cancer screening 2021. Official journal of the American College of Gastroenterology| ACG, 116(3):458–479.
- Siegel et al. (2019) Rebecca L Siegel, Kimberly D Miller, and Ahmedin Jemal. 2019. Cancer statistics, 2019. CA: a cancer journal for clinicians, 69(1):7–34.
- Snoek et al. (2012) Jasper Snoek, Hugo Larochelle, and Ryan P Adams. 2012. Practical bayesian optimization of machine learning algorithms. Advances in neural information processing systems, 25.
- Sundararajan et al. (2017) Mukund Sundararajan, Ankur Taly, and Qiqi Yan. 2017. Axiomatic attribution for deep networks. In International conference on machine learning, pages 3319–3328. PMLR.
- Weischedel et al. (2011) Ralph Weischedel, Sameer Pradhan, Lance Ramshaw, Martha Palmer, Nianwen Xue, Mitchell Marcus, Ann Taylor, Craig Greenberg, Eduard Hovy, Robert Belvin, et al. 2011. Ontonotes release 4.0. LDC2011T03, Philadelphia, Penn.: Linguistic Data Consortium.
- Winawer and Zauber (2002) Sidney J Winawer and Ann G Zauber. 2002. The advanced adenoma as the primary target of screening. Gastrointestinal Endoscopy Clinics, 12(1):1–9.
- Wu et al. (2020) Jialun Wu, Kaiwen Tang, Haichuan Zhang, Chunbao Wang, and Chen Li. 2020. Structured information extraction of pathology reports with attention-based graph convolutional network. In 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 2395–2402. IEEE.
- Yala et al. (2017) Adam Yala, Regina Barzilay, Laura Salama, Molly Griffin, Grace Sollender, Aditya Bardia, Constance Lehman, Julliette M Buckley, Suzanne B Coopey, Fernanda Polubriaginof, et al. 2017. Using machine learning to parse breast pathology reports. Breast cancer research and treatment, 161(2):203–211.
- Zhong et al. (2018) Yizhen Zhong, Luke Rasmussen, Yu Deng, Jennifer Pacheco, Maureen Smith, Justin Starren, Wei-Qi Wei, Peter Speltz, Joshua Denny, Nephi Walton, et al. 2018. Characterizing design patterns of ehr-driven phenotype extraction algorithms. In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1143–1146. IEEE.
Appendix A Appendix
A.1 CRC status annotation criteria
-
•
CRC
-
–
All stages (I-IV)
-
–
-
•
Advanced adenoma (AA)
-
–
Adenoma with carcinoma in situ or high-grade dysplasia, any size
-
–
Adenoma, any villous features, any size
-
–
Adenoma cm in size
-
–
Serrated lesion, cm in size, including sessile serrated adenoma/polyp (SSA/P) with or without cytological dysplasia and hyperplastic polyps (HP) cm
-
–
Traditional serrated adenoma (TSA), any size
-
–
-
•
Non-advanced adenoma (NAA)
-
–
Any number of adenomas, all < 1.0 cm in size, non-advanced
-
–
-
•
Negative (NEG)
-
–
All SSA/P < 1.0 cm and HP < 1.0 cm NOT in sigmoid or rectum
-
–
HP < 1.0 cm in the sigmoid or rectum
-
–
Negative upon histopathological review
-
–
No findings on colonoscopy, no histopathological review
-
–
| Sample Count | NEG | NAA | AA | CRC | Total |
|---|---|---|---|---|---|
| Train+Val set | 1,221 |
482 |
273 |
173 |
2,149 |
| Test set | 515 |
183 |
146 |
75 |
919 |
| Model | Train | Validation | Test | ||||||
|---|---|---|---|---|---|---|---|---|---|
| F1 | precision | recall | F1 | precision | recall | F1 | precision | recall | |
| BoW | 1.000 | 1.000 | 1.000 | 0.909 | 0.926 | 0.897 | 0.839 | 0.845 | 0.838 |
| CNN | 0.998 | 0.998 | 1.000 | 0.920 | 0.932 | 0.911 | 0.883 | 0.886 | 0.882 |
| BioBERT | 0.960 | 0.955 | 0.965 | 0.946 | 0.946 | 0.946 | 0.914 | 0.904 | 0.925 |
| BioBERT+Lesion Size | 1.000 | 1.000 | 1.000 | 0.921 | 0.921 | 0.927 | 0.923 | 0.920 | 0.930 |
| Hyperparameters | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BoW |
|
|||||||||||||
| CNN |
|
|||||||||||||
| BioBERT |
|