JOINT OPTIMIZATION OF PIECEWISE LINEAR ENSEMBLES
Abstract
Tree ensembles achieve state-of-the-art performance on numerous prediction tasks. We propose Joint Optimization of Piecewise Linear Ensembles (JOPLEn), which jointly fits piecewise linear models at all leaf nodes of an existing tree ensemble. In addition to enhancing the ensemble expressiveness, JOPLEn allows several common penalties, including sparsity-promoting and subspace-norms, to be applied to nonlinear prediction. For example, JOPLEn with a nuclear norm penalty learns subspace-aligned functions. Additionally, JOPLEn (combined with a Dirty LASSO penalty) is an effective feature selection method for nonlinear prediction in multitask learning. Finally, we demonstrate the performance of JOPLEn on 153 regression and classification datasets and with a variety of penalties. JOPLEn leads to improved prediction performance relative to not only standard random forest and boosted tree ensembles, but also other methods for enhancing tree ensembles.
While preparing the PyPI package for general release, we found minor bugs in the penalty gradient computation and the validation set preprocessing that affected JOPLEn with a Laplacian + Frobenius-norm penalty and JOPLEn/GR with a CatBoost partitioner, respectively. Fixing these bugs provides the updated results shown in Figure 1 and Section 3.1. The conclusions of the paper remain the same.
Index Terms— Joint, global, optimization, refinement, feature selection, subspace, ensemble, linear
1 Introduction
Ensemble methods combine multiple prediction functions into a single prediction function. A canonical example is a tree ensemble, where each tree “partitions” the feature space, and each leaf is a “cell” of the partition containing a simple (e.g., constant) model. Although neural networks (NN) have recently triumphed on structured data, they have struggled to outperform tree ensembles, such as gradient boosting (GB), across diverse tabular (i.e., table-formatted) datasets [1].
Despite the longstanding success of tree ensembles, standard implementations are still limited, either by the piecewise constant fits at each cell, or by the greedy, suboptimal way in which the ensembles are trained. To address these limitations, several improvement strategies have been proposed. FASTEL uses backpropagation to optimize the parameters of an ensemble of smooth, piecewise constant trees [2]. Global refinement (GR) jointly refits all constant leaves of a tree ensemble after first running a standard training algorithm, such a random forests or gradient boosting [3]. Partition-wise Linear Models also perform joint optimization, but learn linear functions on axis-aligned and equally-spaced step functions [4]. Linear Forests (LF) increase model expressiveness by replacing constant leaves with linear models [5].
Because tree ensembles are nonlinear and greedily constructed, it has also been challenging to incorporate structure-promoting penalties through joint optimization. RF feature selection typically relies on a heuristic related to the total impurity decrease associated to each feature. This approach may underselect correlated features, and requires further heuristics to extend to the multitask setting. GB feature selection uses a greedy approximation of the norm, which penalizes the addition of new features to each subsequent tree [6]. BoUTS is a multitask extension that selects “universal” features (important for all tasks) and “task-specific” features by selecting features that maximize the minimum impurity decrease across all tasks [7]. As such examples demonstrate, the incorporation of feature sparsity and similar structural objectives into tree ensembles has been limited by the greedy nature of ensemble construction.
We propose Joint Optimization of Piecewise Linear Ensembles (JOPLEn), an extension of global refinement (GR) that is applied to ensembles of piecewise linear functions. JOPLEn both ameliorates greedy optimization and increases model flexibility by jointly optimizing a hyperplane in each cell (e.g., leaf) in each partition (e.g., tree) of an ensemble. Besides improving performance, and unlike prior tree ensembles, JOPLEn is compatible with many standard structure-promoting regularizers, which provides a simple way to incorporate sparsity, subspace structure, and smoothness into nonlinear prediction. We demonstrate this capability on 153 real-world and synthetic datasets; JOPLEn frequently outperforms existing methods for regression, binary classification, and multitask feature selection, including GB, RF, LF, CatBoost, soft decision trees (FASTEL), and NNs. Finally, we provide a GPU-accelerated implementation that is extensible to new loss functions, partitions, and regularizers.
2 Methodology
We begin by describing JOPLEn in the single task setting, and subsequently extend it to multitask learning.
2.1 Single-task JOPLEn
Let be the feature space and the output space. For regression, , and for binary classification, . Let be a training dataset where , , and . A model class is a set of functions . Given , the goal of single-task supervised learning is to find an that accurately maps feature vectors to outputs. For regression and binary classification, predictions are made using and , respectively.
JOPLEn is an instance of (regularized) empirical risk minimization (ERM). In ERM, a prediction function is a solution of
| (1) |
where is a regularization term (penalty), and is a loss function. We take to be the squared error loss for regression, and the logistic loss for binary classification.
We focus on the setting where is an additive ensemble , where is defined piecewise on a partition (e.g., a decision tree), and there are partitions. The partitions are fixed in advance, and may be obtained by running a standard implementation of a tree ensemble model. Further, we assume that . Here is the number of “cells” in the partition (e.g., tree), is a weight vector, is a bias term, and indicates whether data point is in cell of partition [3] (e.g., indicating the decision regions of a tree). Then, we denote a piecewise linear ensemble as
| (2) |
where is the total number of cells, is the matrix of all weight vectors, is a vector of all bias terms, and is the component of associated with the th cell of partition . JOPLEn is then defined by the solution of
| (3) |
where is a regularization term (penalty). The notation indicates that may depend on , but that only and will be penalized.
JOPLEn has attractive optimization properties by construction. For example, a convex loss and regularization term will render the overall objective convex. We optimize JOPLEn using Nesterov’s accelerated gradient method, and we use a proximal operator for any non-smooth penalties.
JOPLEn easily incorporates existing penalties, and provides a straightforward framework for extending well-known penalties for linear models to a nonlinear setting. Notable examples include sparsity-promoting matrix norms (e.g., -norms [8, 9] and GrOWL [10]) and subspace norms (nuclear, Ky Fan, and Schatten -norms) [11]. We demonstrate two such standard penalties: -norms and the nuclear norm.
2.1.1 Single-task -norm
Because we use linear leaf nodes, we can use sparsity-promoting penalties to preform feature selection. To learn a consistent sparsity pattern across all linear terms, we realize as an sparsity-promoting group norm (for ), which leads to a row-sparse solution for . Specifically,
| (4) |
where is the th component of the vector . Concretely, we use the norm, which has a straightforward proximal operator [12, Eq. 6.8, p. 136].
2.1.2 Single-task nuclear norm
Given a singular value decomposition for the conjugate transpose , the nuclear norm is defined as
| (5) |
When applied to JOPLEn’s weights, the nuclear norm penalty empirically functions similarly to a group-norm penalty. While group-norms promote sparse solutions in an axis-aligned subspace, the nuclear norm apparently promotes “sparse” solutions along the “axes” of a data-defined subspace. This norm also has a straightforward proximal operator using singular value thresholding [12, Eq. 6.13].
2.2 Multitask JOPLEn
Suppose we have datasets indexed by : . Multitask JOPLEn is nearly identical to the single-task case, but includes an additional sum over tasks. Let for a task-specific number of cells , , and be the -th task’s weight matrix, bias vector, partitioning function, and relative weight. For notational simplicity, define , , and for the transpose . Then multitask JOPLEn is defined as
| (6) |
and can be optimized using the same approach as in the single-task setting.
2.2.1 Dirty LASSO
JOPLEn with an extended Dirty LASSO (DL) [13] penalty performs feature selection for nonlinear multitask learning. Suppose that we know a priori that some features are important for all tasks, some features are important for only some tasks, and all other features are irrelevant. Then we can apply a JOPLEn extension of DL to perform “common” and “task-specific” feature selection.
DL decomposes the weight matrix , encouraging to be row-sparse (common features) and each to be individually row-sparse (task-specific features), with potentially different sparsity patterns for each . Given penalty weights , JOPLEn DL is the solution of
| (7) |
a combination of LASSO and Group LASSO penalties [13].
2.3 Laplacian regularization
The naïve use of piecewise-linear functions may lead to pathological discontinuities at cell boundaries. Thus, we utilize graph Laplacian regularization [14] to force nearby points to have similar values. Using the standard graph Laplacian,
| (8) |
where is the model prediction for a feature vector and is a distance-based kernel. In this paper, is a Gaussian radial basis function. Laplacian regularization can be naïvely applied to each task in the multitask setting as well.
3 Experiments
We evaluate JOPLEn in multiple regression and classification settings with several regularization terms.
3.1 Single-task regression and binary classification
We evaluate JOPLEn’s predictive performance on the “Penn Machine Learning Benchmark” (PMLB) of 284 regression, binary classification, and multiclass classification tasks [15]. For simplicity, we focus on regression and binary classification. Some methods (e.g., GB, JOPLEn) only handle real-valued features, so we drop all other features for such methods. We ignore datasets that have no real-valued features, or are too large for our GPU. This leaves 90 regression and 60 binary classification datasets. For each dataset, we perform a random 0.8/0.1/0.1 train/validation/test split.
To facilitate comparisons with previous methods [3], we jointly optimize partitions created using Gradient Boosted trees [16], Random Forests [17], and CatBoost [18]. We also evaluate JOPLEn with random Voronoi ensembles (Vor). Each partition in a Voronoi ensemble is created by sampling data points uniformly at random and creating Voronoi cells from these data points. This is to provide context for the performance of JOPLEn in Section 3.3 and Figure 3 b).
To demonstrate JOPLEn’s efficacy, we benchmark several alternative models: Gradient Boosted trees (GB), Random Forests (RF) [19], Linear Forests (LF) [20, 5], and CatBoost (CB) [18]; Linear/Logistic Ridge Regression [19]; a feedforward neural network (NN) [21] and a NN for tabular data (TabNet) [22, 23]; a differentiable tree ensemble (FASTEL) [2]; and global refinement (GR) [3]. GB, RF, LF, and CB are baseline ensembles for demonstrating the performance improvement from joint optimization. CB utilizes categorical features and corrects a bias in GB’s gradient step [18]. FASTEL and TabNet provide a comparison between joint optimization, fully-differentiable ensembles, and tabular deep learning. We evaluate CatBoost both with and without (NC) categorical features. We exclude FASTEL from the classification experiments because the code does not include classification losses. Global refinement (GR) jointly optimizes piecewise-constant tree ensembles. We exclude the global pruning step from [3] to more clearly demonstrate the direct performance improvement of JOPLEn’s piecewise-linear approach. These approaches provide a thorough comparison between JOPLEn and existing methods.
As in [3], we use the squared Frobenius norm of leaf nodes for both piecewise-constant and piecewise-linear ensembles. We also evaluate a combined Laplacian-Frobenius penalty for piecewise-linear ensembles (Section 2.3).
We optimize model hyperparameters using Ax [24], which combines Bayesian and bandit optimization for continuous and discrete parameters. Ax uses 50 training/validation trials to model the validation set’s empirical risk as a function of hyperparameters, and we use the optimal hyperparameters to evaluate model performance on the test set. Hyperparameter ranges are documented in our code.
To facilitate plotting, we normalize the mean squared error (MSE) of each regression dataset by the MSE achieved by predicting the training mean and call this the “normalized” MSE. The 0/1 loss does not require normalization.
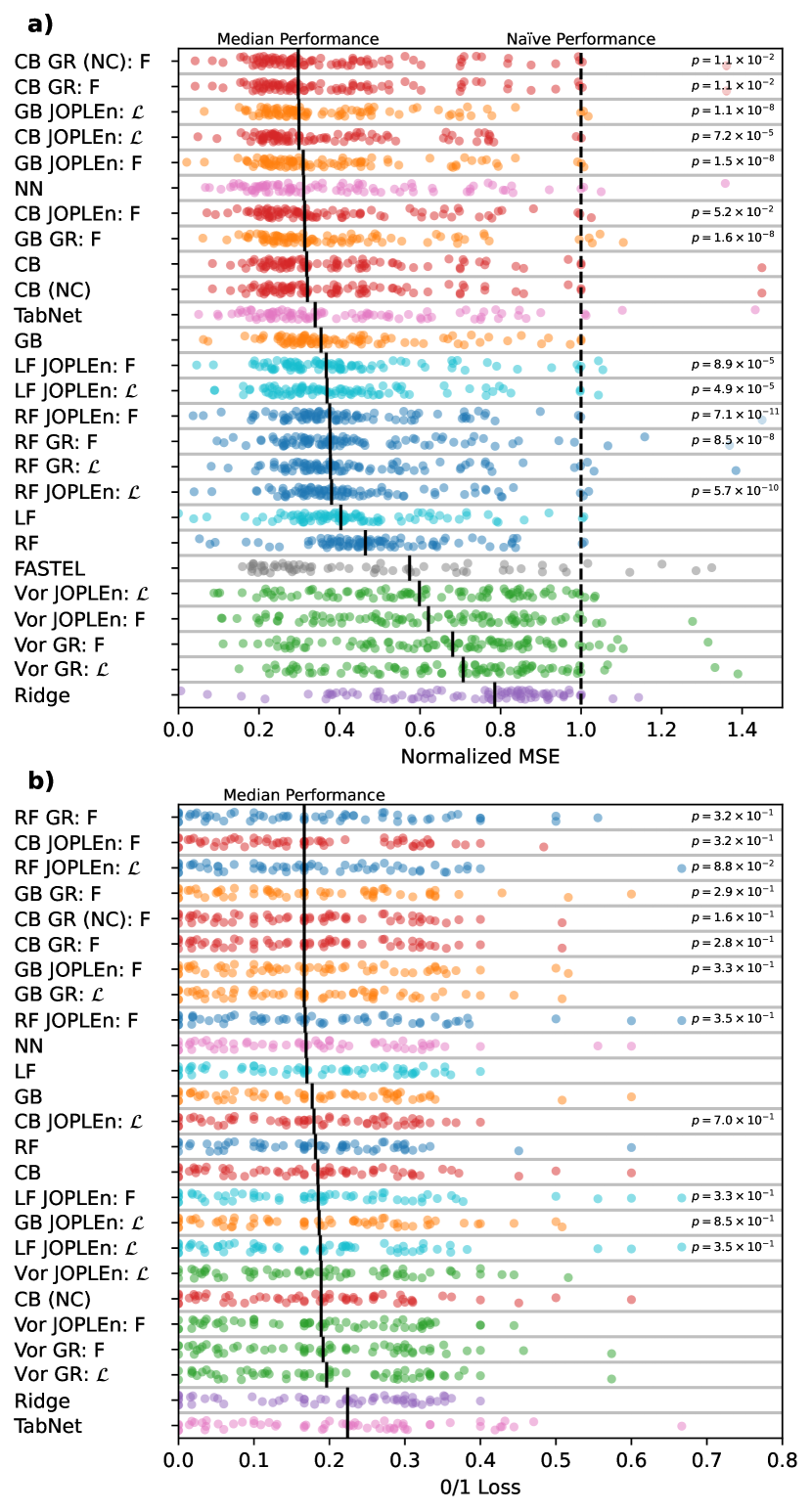
Similar to [3], Figure 1 a) shows that JOPLEn and GR significantly improve the regression performance of GB and RF, as determined by a one-sided Wilcoxon signed-rank test. JOPLEn also significantly improves LF and CB. Indeed, JOPLEn CB outperforms all regression approaches including CB (), despite dropping all categorical features. Based on median performance, CB GR (NC), CB GR, and GB JOPLEn appear to outperform JOPLEn CB . However, JOPLEn CB outperforms them with , , and due to their heavier tail performance.
Although JOPLEn also improves performance for binary classification, the difference is less pronounced ( for JOPLEn RF ). This decreased significance may be caused by the sample size (60 vs. 90 datasets), or the fact that the 0/1 loss only considers the sign of the prediction.
Interestingly, we observe that the partitioning method may significantly affect performance. In the regression setting, CB, GB, LF, RF, and Voronoi model performances rank in this order. However, there is no significant difference in the classification setting. We suspect that there may be other partitioning methods that lead to further performance increases.
Finally, we note that CB JOPLEn outperforms the NNs and FASTEL in general, achieving -values of and for TabNet and the feedforward network. The NNs and FASTEL perform well on many datasets, but have a high median loss because some datasets perform quite poorly. This is particularly true for FASTEL.
3.2 Single-task nuclear norm
In Figure 2 we demonstrate the effect of the nuclear norm on synthetic data. We draw 10,000 samples (100 are training data) uniformly on the square interval , and define where for an input feature and . Here, we use random Voronoi ensembles and fix the number of partitions and cells to simplify visualization, and manually tune the penalty weights.
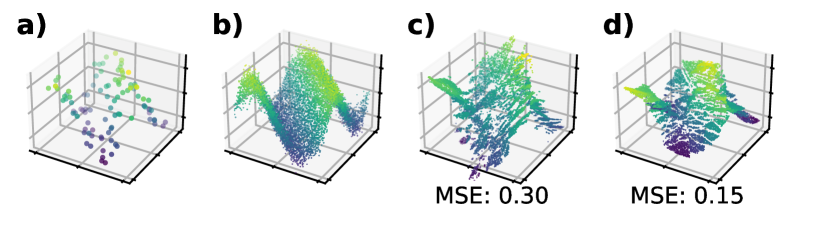
Notably, the nuclear norm causes the model to align its piecewise linear functions along the diagonal, forming a “consensus” across all linear models. By contrast, the Frobenius norm penalty allows each leaf to be optimized independently, and thus suffers from degraded performance.
3.3 Multitask Dirty LASSO
Next, we show that JOPLEn Dirty LASSO (DL) can achieve superior sparsity to the DL [13].
We selected two multitask regression datasets from the literature: NanoChem [7] and SARCOS [25]. NanoChem is a group of 7 small molecule, nanoparticle, and protein datasets for evaluating multitask feature selection performance [7]. These datasets have 1,205 features and 127 to 11,079 samples. The response variables are small molecule boiling point (1), Henry’s constant (2), logP (3) and melting point (4); nanoparticle logP (5) and zeta potential (6); and protein solubility (7). SARCOS is a 7-task dataset that models the dynamics of a robotic arm [25], with 27 features and 48,933 samples for each task. These datasets were also split into train, validation, and test sets using a 0.8/0.1/0.1 ratio.
Using JOPLEn DL with tree ensembles will provide biased sparsity estimates; features are used to create each tree (not penalized) and then to train each linear leaf (penalized). In this case, simply analyzing the leaf weights will underestimate the number of features used by the JOPLEn model. We avoid this issue by using random Voronoi ensembles.
All penalties were manually tuned so that all methods achieve a similar test loss for each task. Not all datasets are equally challenging, so we manually tune the parameter for JOPLEn DL and BoUTS. No equivalent parameters exist in the community implementation of DL [26].
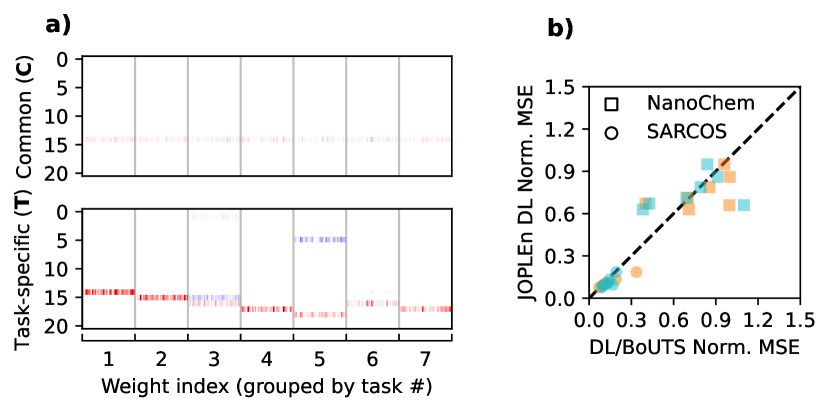
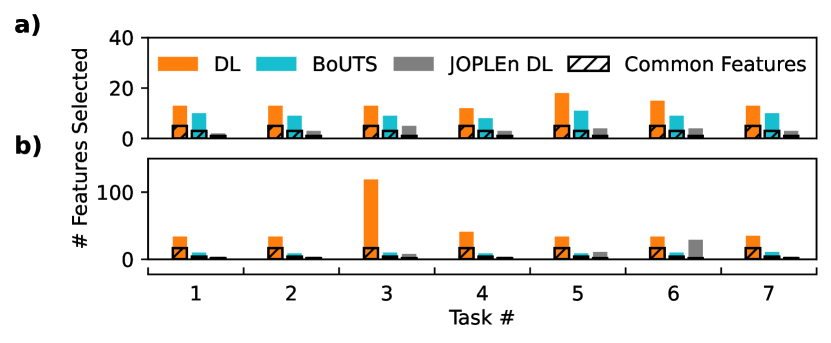
We find that JOPLEn DL and BoUTS select significantly sparser feature sets than DL does, with JOPLEn providing the sparsest sets. Figure 3 a) shows that joint optimization shares penalties across ensemble terms, providing structured sparsity. Further, JOPLEn selects far fewer features than DL (Fig. 4) while achieving similar or superior performance on all but one task (small molecule logP (3), Fig. 3 b)). This is likely because of the significant disparity in the number of features selected for this task (102 for DL, vs. 6 for JOPLEn).
4 Conclusions
The JOPLEn framework enables many existing penalties for linear methods (e.g., single- and multitask feature selection) to be incorporated into nonlinear methods.
We find that JOPLEn beats global refinement (GR) and significantly outperforms other state-of-the-art methods on real-world and synthetic datasets. We demonstrate that Laplacian; Frobenius, nuclear, -norm; and Dirty LASSO regulation have straightforward extensions using JOPLEn and improve model performance. Empirically, JOPLEn shines when the response variable is a nonlinear function of input features with structured sparsity, such as multitask feature selection. Such settings cannot be modeled using linear approaches such as LASSO, and JOPLEn achieves similar or higher feature sparsity and performance than BoUTS (Figure 3 b)) while using suboptimal partitions (Figure 1). Future work may use BoUTS as a partitioner for JOPLEn to improve performance while maintaining a high degree of sparsity. Combining JOPLEn with global pruning, developing new partitioning methods, and providing support for categorical features may also lead to further improvements. Overall, these results suggest that JOPLEn is a promising approach to improving performance on tabular datasets.
We anticipate that JOPLEn will improve regression and classification performance across many fields. Additionally, we expect that JOPLEn’s piecewise linear formulation will lead to improved interpretability through increased feature sparsity. Finally, because JOPLEn allows the straightforward transfer of linear penalties to the nonlinear setting, we anticipate that it will greatly simplify the implementation of nonlinear penalized optimization problems (e.g., subspace-aligned nonlinear regression via the nuclear norm, Figure 2).
5 Code availability
A JAX (CPU and GPU compatible) version of JOPLEn is available on PyPI as joplen with linked source code. The data, JOPLEn implementation, and evaluation/plotting code for this paper is available at https://gitlab.eecs.umich.edu/mattrmd-public/joplen-repositories/joplen-mlsp2024.
References
- [1] Ravid Shwartz-Ziv and Amitai Armon, “Tabular data: Deep learning is not all you need,” Inform. Fusion, vol. 81, 2022.
- [2] Shibal Ibrahim, Hussein Hazimeh, and Rahul Mazumder, “Flexible modeling and multitask learning using differentiable tree ensembles,” in SIGKDD, 2022.
- [3] Shaoqing Ren, Xudong Cao, Yichen Wei, and Jian Sun, “Global refinement of random forest,” in IEEE CVPR, 2015.
- [4] Hidekazu Oiwa and Ryohei Fujimaki, “Partition-wise linear models,” in NeurIPS, 2014.
- [5] Juan J. Rodríguez, César García-Osorio, Jesús Maudes, and José Francisco Díez-Pastor, “An experimental study on ensembles of functional trees,” in Lect. Notes Comput. Sc., 2010, vol. 5997.
- [6] Zhixiang Xu, Gao Huang, Kilian Q. Weinberger, and Alice X. Zheng, “Gradient boosted feature selection,” in SIGKDD, 2014.
- [7] Matt Raymond, Jacob Charles Saldinger, Paolo Elvati, Clayton Scott, and Angela Violi, “Universal feature selection for simultaneous interpretability of multitask datasets,” arXiv, 2024.
- [8] Guillaume Obozinski, Ben Taskar, and Michael Jordan, “Multi-task feature selection,” Statistics Department, UC Berkeley, Tech. Rep, vol. 2, no. 2.2, 2006.
- [9] Yaohua Hu, Chong Li, Kaiwen Meng, Jing Qin, and Xiaoqi Yang, “Group sparse optimization via regularization,” JMLR, vol. 18, no. 30, 2017.
- [10] Urvashi Oswal, Christopher Cox, Matthew Lambon-Ralph, Timothy Rogers, and Robert Nowak, “Representational similarity learning with application to brain networks,” in ICML, 2016.
- [11] Jeffrey A. Fessler and Raj Rao Nadakuditi, Linear Algebra for Data Science, Machine Learning, and Signal Processing, Cambridge University Press, 2024.
- [12] Neal Parikh and Stephen Boyd, “Proximal algorithms,” Found. Trends Optim., vol. 1, no. 3, Jan 2014.
- [13] Ali Jalali, Sujay Sanghavi, Chao Ruan, and Pradeep Ravikumar, “A dirty model for multi-task learning,” in NeurIPS, 2010.
- [14] Xiaojin Zhu, Zoubin Ghahramani, and John Lafferty, “Semi-supervised learning using gaussian fields and harmonic functions,” in ICML, 2003.
- [15] Joseph D Romano, Trang T Le, William La Cava, et al., “PMLB v1.0: an open source dataset collection for benchmarking machine learning methods,” arXiv, 2021.
- [16] Jerome H. Friedman, “Greedy function approximation: A gradient boosting machine,” Ann. Stat., vol. 29, no. 5, 1999.
- [17] Leo Breiman, “Bagging predictors,” Mach. Learn., vol. 24, no. 2, Aug 1996.
- [18] Liudmila Prokhorenkova, Gleb Gusev, Aleksandr Vorobev, Anna Veronika Dorogush, and Andrey Gulin, “CatBoost: unbiased boosting with categorical features,” in NeurIPS, 2018, vol. 31.
- [19] F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, and E. Duchesnay, “SciKit-Learn: Machine learning in Python,” JMLR, vol. 12, 2011.
- [20] Marco Cerliani, “linear-tree,” PyPI, 2022, version 0.3.5.
- [21] Jeffrey Dean, Rajat Monga, et al., “TensorFlow: Large-scale machine learning on heterogeneous systems,” 2015, Software available from tensorflow.org.
- [22] Sercan O. Arik and Tomas Pfister, “TabNet: Attentive interpretable tabular learning,” arXiv, 2020.
- [23] Eduardo Carvalho Pinto and Hartorn Fischman, Sébastien, “Pytorch TabNet,” PyPI, 2019, version 4.1.0.
- [24] Eytan Bakshy, Lili Dworkin, Brian Karrer, Konstantin Kashin, Ben Letham, Ashwin Murthy, and Shaun Singh, “AE: A domain-agnostic platform for adaptive experimentation,” in NeurIPS Systems for ML Workshop, 2018.
- [25] Sethu Vijayakumar, Aaron D’Souza, and Stefan Schaal, “Incremental Online Learning in High Dimensions,” Neural Comput., vol. 17, 12 2005.
- [26] Hicham Janati, “MuTaR: Multi-task regression in Python,” 2019, Github hash: b682ba9.