Leveraging SAM for Single-Source Domain Generalization in Medical Image Segmentation
Abstract
Domain Generalization (DG) aims to reduce domain shifts between domains to achieve promising performance on the unseen target domain, which has been widely practiced in medical image segmentation. Single-source domain generalization (SDG) is the most challenging setting that trains on only one source domain. Although existing methods have made considerable progress on SDG of medical image segmentation, the performances are still far from the applicable standards when faced with a relatively large domain shift. In this paper, we leverage the Segment Anything Model (SAM) to SDG to greatly improve the ability of generalization. Specifically, we introduce a parallel framework, the source images are sent into the SAM module and normal segmentation module respectively. To reduce the calculation resources, we apply a merging strategy before sending images to the SAM module. We extract the bounding boxes from the segmentation module and send the refined version as prompts to the SAM module. We evaluate our model on a classic DG dataset and achieve competitive results compared to other state-of-the-art DG methods. Furthermore, We conducted a series of ablation experiments to prove the effectiveness of the proposed method. The code is publicly available at https://github.com/SARIHUST/SAMMed.
1 Introduction
Medical image segmentation plays a crucial role in various healthcare applications, enabling accurate delineation and analysis of anatomical structures or abnormalities within medical images. However, achieving robust and accurate segmentation in medical imaging poses unique challenges due to the complexity and variability of anatomical structures, imaging modalities, noise, and artifacts. In the field of medical image segmentation, domain generalization becomes particularly important due to the significant variations in image styles and characteristics across different medical equipment. Different manufacturers, models, and settings of medical imaging devices can result in distinct image styles even for the same type of target. This inherent diversity makes it essential to develop segmentation algorithms that can effectively generalize across different medical domains, ensuring reliable and accurate segmentation results across a wide range of imaging scenarios.
Since its introduction, the Segment Anything Model (SAM) has shown outstanding performance across a wide spectrum of image segmentation tasks. Thanks to its remarkable flexibility in segmenting objects using diverse prompts, including points, bounding boxes, and text description, SAM possesses the extraordinary capability to perform segmentation tasks across a wide range of scenarios without requiring the burden of additional annotation and training, which makes it well suited for domain generalization tasks.
However, in fields where precision is of utmost importance, such as the challenging field of Medical Image Processing, it’s worth noting that SAM may not provide the most satisfactory outcomes. Furthermore, SAM relies heavily on the usage of high-quality bounding box prompts to achieve accurate and reliable segmentation results. Unlike regular images, in the context of medical images, the incorporation of such bounding boxes requires a significant level of expertise and domain-specific knowledge. Therefore, while SAM exhibits impressive capabilities in various scenarios, its effectiveness in medical image processing tasks should be evaluated with careful consideration, recognizing the need for further improvements to meet the specific requirements and challenges of the domain.
In contrast, conventional approaches to medical image segmentation do not necessitate the provision of bounding boxes; rather, they heavily rely on meticulously crafted network architectures, which inherently limits their applicability in tasks involving domain generalization. This limitation arises due to the intricate design and manual engineering of these methods, which restricts their ability to adapt and generalize across diverse medical imaging datasets and scenarios.
In order to harness the strengths of both SAM and specific domain knowledge, we now propose a new fine-tuning paradigm. To address the bounding box provision problem of SAM, our approach utilizes a traditional segmentation network to predict coarse masks and then generate refined bounding boxes. We then fine-tune SAM’s lightweight mask decoder to generate final prediction masks.
Our main contributions are as follows: (1) We propose a novel fine-tuning paradigm to leverage SAM for single-sourced domain generalization tasks in medical image segmentation. (2) To fully leverage SAM’s image encoder and prompt encoder, we employed image merging and mask filtering techniques to reduce the inference time and enhance overall mask predicting performance. (3) Our approach achieves state-of-the-art performance on the Prostate dataset.
2 Related Works
2.1 Segment Anything Model
The Segment Anything Model (SAM) proposed by Kirillov et al. (2023) stands out as a groundbreaking foundational model that introduces the concept of promptability, enabling robust generalization across numerous domains. Trained on the large SA-1B dataset, SAM has demonstrated exceptional performance in zero-shot segmentation of natural images, showcasing its versatility and adaptability. However, when it comes to medical image segmentation, SAM’s original model encounters limitations due to its lack of domain-specific knowledge in the realm of medical imaging as demonstrated in Zhang & Jiao (2023).
To address this challenge, previous works have explored various strategies to enhance SAM’s performance in medical image segmentation tasks. One such approach involves fine-tuning SAM using medical images, as demonstrated in projects like MedSAM(Ma & Wang, 2023) and MSA(Wu et al., 2023). Some of the work like Skinsam(Hu et al., 2023) and SAMUS(Lin et al., 2023) applied SAM to more specific segmentation tasks including ultrasound, 3d images, or skin cancer. They aim to impart SAM with a better understanding of medical images’ unique characteristics and intricacies, enabling it to produce more accurate and reliable segmentation results. Another promising solution is the utilization of auto-prompting techniques, which have shown promising improvements in SAM’s performance across different domains in projects like AutoSAM(Shaharabany et al., 2023) and AutoSAM Adapter(Li et al., 2023).
Despite these advancements, it is important to note that the current performance achieved by these approaches may not be sufficient for clinical applications that demand higher levels of accuracy and reliability. Additionally, it is worth highlighting that there is no noticeable research focused on the broader aspect of generalization across domains in the context of SAM. While SAM has demonstrated impressive generalization capabilities in various scenarios, there remains a need to explore and develop techniques that enable SAM to effectively generalize across diverse domains, including medical imaging. By addressing this research gap, we can unlock SAM’s full potential in tackling the challenges associated with domain generalization and further elevate its applicability in real-world medical image segmentation tasks.
2.2 Domain Generalization & Single-Source Domain Generalization
Image segmentation is critical for computer-aided diagnosis and treatment planning. However, the effectiveness of deep network models trained for specific domains can be hindered when confronted with new, unseen target domains due to unpredictable domain shifts. This challenge has spurred significant interest in the field of domain generalization (DG), which aims to develop models that can generalize well across diverse domains, even those that have not been encountered during training. Several recent works have achieved good performance in medical image segmentation, by applying data augmentation and learning domain-invariant features. So far, some works have applied domain generalization to medical image segmentation. For instance, Liu et al. (2021) proposed a method based on meta-learning by encouraging the shape compactness and shape smoothness for prostate segmentation. Wang et al. (2020) developed a Domain Knowledge Pool to learn and memorize the prior information extracted from multi-source domains, which helps to augment the original image. Hu et al. (2022) designed a domain adaptive convolution module and content adaptive convolution module and incorporated both into an encoder-decoder backbone. Zhou et al. (2022) introduced a domain-specific restoration module for regularization and a Random Amplitude Mixup module with low-level frequency knowledge.
Among the various scenarios in DG, single-source domain generalization presents a distinct challenge. Unlike multi-source domain generalization, which benefits from multiple source domains, single-source DG aims to train a predictive model that is both robust and generalizable using only a single source domain. This poses an extreme lack of data, making it more difficult to achieve reliable and accurate segmentation results. Recent research efforts have primarily focused on addressing this challenge through innovative techniques such as data augmentation and the integration of multi-source DG techniques like adversarial training and contrastive learning.Ouyang et al. (2022) proposed a simple causality-inspired data augmentation approach to expose a segmentation model to synthesized domain-shifted training examples. Su et al. (2023) proposed a location-scale augmentation strategy, that engages the inherent class-level information and enjoys a general form of augmentation. Xu et al. (2022) proposed a mutual information regularizer to enforce the semantic consistency between images from the synthetic domains and estimated the consistency by patch-level contrastive learning. However, the outcomes obtained thus far have been considerably unsatisfying.
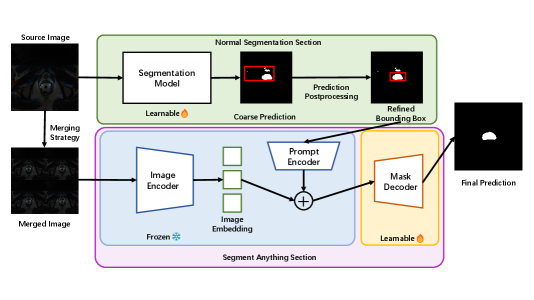
3 Methods
3.1 Architecture Overview
The overview of our methods architecture is shown in Figure 1. Our approach compromises two primary stages. In the first stage, we utilize a traditional segmentation model to generate coarse prediction masks. We then apply a mask-filtering module to produce refined bounding boxes, which are used in the second stage to fine-tune SAM. Each stage and its main components are described in detail in the following sections.
3.2 Segmentation Backbone
In the first stage, we train a Resnet backbone as a segmentation network to produce coarse prediction masks. The segmentation network is given input of and outputs , where and denote the source image, ground truth masks, and coarse predictions respectively. Resnet was selected as the backbone network due to its widespread adoption and established reputation in the domain of medical image segmentation.
During the training phase, the network is exclusively trained using data from a single domain. In the subsequent testing phase, the trained network is directly employed on unfamiliar target domains without any additional modifications.
3.3 Mask-Filtering Module
To ensure the production of high-quality bounding boxes, we acknowledge the presence of considerable noise within the coarse masks initially predicted by Resnet. This noise has the potential to significantly degrade the quality of the resulting bounding boxes, which would lead to unpromising mask prediction results from SAM.
To address this issue, we introduce a mask-filtering module that effectively eliminates noise and retains the largest continuous mask, thereby improving the overall quality of the generated bounding boxes. The underlying concept of this mask-filtering module is straightforward yet impactful. Despite the presence of considerable noise in the output of the segmentation network, it is highly probable that the network accurately identifies the largest region corresponding to the target object. Leveraging this insight, we employ a breadth-first search (BFS) algorithm to identify the largest connected component of the mask, discarding all other components. This refinement process yields improved predictions and facilitates the generation of more precise bounding boxes.
While implementing this module, we observed that the computational speed of the algorithm is significantly influenced by the size of the input image. Considering that the main objective of this step is to eliminate smaller components in the coarse prediction masks, preserving the original size is unnecessary. We devised a strategy to expedite this process by resizing the initial coarse prediction masks to a smaller dimension , (e.g. from to ), before executing the algorithm. Subsequently, to restore the original size of the masks, we upscale the corresponding bounding box ’s parameters by the corresponding factor (e.g. 4 for to ). By adopting this approach, we achieve a balance between computational efficiency and the desired accuracy of the bounding boxes.
3.4 SAM Section
Following the acquisition of refined bounding boxes, we employed a fine-tuning strategy on SAM, similar to the approach used in MedSAM. During this process, we exclusively fine-tuned SAM’s lightweight mask decoder, while keeping the parameters of the image encoder and prompt encoder fixed throughout training.
However, it is worth noting that MedSAM resizes all images to a standardized dimension of to accommodate SAM’s embedding module. Unfortunately, this resizing operation can incur unnecessary computational overhead and impede training speed in certain situations.
In contrast, to fully exploit the capabilities of SAM’s robust image encoder and its multi-box prediction functionality, we adopted a straightforward concatenation method. Specifically, we merged four individual images to create a larger composite image with dimensions of . Consequently, the associated bounding boxes and ground truth masks were adjusted accordingly. This merging approach enables us to harness the benefits of SAM’s pre-trained embedding modules within its image encoder, while simultaneously expediting the training and testing processes.
Similar to section 3.2, during the training phase, SAM is only fine-tuned using only the data from the same single source domain, while tested on all other target domains.
4 Experiments
4.1 Experiment Setting
Dataset In this study, we assess the performance of our methodology using the publicly available Prostate dataset, which is a diverse collection of T2-weighted MRI prostate images and corresponding masks, spanning six distinct domains.
Evaluation Metric We employ a widely adopted evaluation metric in medical image segmentation domains - the Dice coefficient (Dice) metric. The Dice coefficient provides a quantitative measure of the quality of segmentation results, with higher values indicating better segmentation performance.
Implementation Details To mitigate the impact of noise in the coarse predictions, we established a higher confidence threshold of for the mask predictions generated by the Resnet backbone. Additionally, considering the superior segmentation performance and increased robustness of the SAM model, we set a lower confidence threshold of for SAM’s predictions. In the training process, we use an Adam solver with a base learning rate of 0.0001 and fine-tune SAM for 200 epochs on each domain of the prostate dataset with a batch size of 16.
4.2 Evaluation on Prostate
We report the performance of our approach on the Prostate dataset in cross-site evaluation, as listed in Table 1. Our approach accomplishes the highest Dice score of , outperforming all previous methods by a large margin. Compared with the former best ACSDG results, our result is higher (absolute) and better (relative).
It is also worth noting that although the previous state-of-the-art methods demonstrated impressive performance in certain domains, they exhibited limitations and shortcomings when applied to other domains. In contrast, our method achieves the best performance in all 6 domains, demonstrating a stronger domain generalization ability.
| Method | A | B | C | D | E | F | AVG |
|---|---|---|---|---|---|---|---|
| ERM | 71.81 | 65.56 | 43.98 | 71.97 | 48.39 | 37.82 | 56.59 |
| Cutout DeVries & Taylor (2017) | 78.36 | 69.08 | 63.45 | 66.39 | 61.88 | 60.19 | 66.56 |
| RSC Huang et al. (2020) | 72.81 | 70.18 | 49.18 | 74.11 | 54.73 | 43.69 | 60.78 |
| MixStyle Zhou et al. (2021) | 73.24 | 58.06 | 44.75 | 66.78 | 49.81 | 49.73 | 57.06 |
| AdvBias Chen et al. (2020) | 78.15 | 62.24 | 54.73 | 72.65 | 53.14 | 51.00 | 61.98 |
| RandConv Xu et al. (2020) | 77.28 | 60.77 | 53.54 | 66.21 | 52.12 | 36.52 | 57.74 |
| CSIDG Ouyang et al. (2022) | 82.14 | 67.21 | 59.11 | 73.16 | 67.38 | 73.23 | 70.37 |
| ACSDG Xu et al. (2022) | - | - | - | - | - | - | 71.42 |
| Ours | 84.08 | 77.29 | 73.98 | 82.40 | 80.47 | 79.04 | 79.54 |
| Improvements | +1.94 | +7.11 | +10.53 | +8.29 | +13.09 | +5.81 | +8.12 |
| Method | A to Rest | B to Rest | C to Rest | D to Rest | E to Rest | F to Rest | Average |
|---|---|---|---|---|---|---|---|
| 0.8408 | 0.7729 | 0.7398 | 0.8240 | 0.8047 | 0.7904 | 0.7954 | |
| 0.8303 | 0.7681 | 0.7288 | 0.8159 | 0.7906 | 0.7929 | 0.7878 | |
| 0.8100 | 0.7577 | 0.7006 | 0.7975 | 0.7786 | 0.7877 | 0.7720 |
4.3 Ablation Studies
We conducted a series of ablation experiments on the Prostate dataset to analyze the design and parameter selection in our method.
Ablation on the confidence threshold of SAM We used different threshold parameters to filter the final prediction of SAM. The performance varies as shown in Table 2. SAM showcases exceptional precision in mask prediction, achieving highly accurate overall prediction results even when the confidence score exceeds only 0.5. However, when a higher confidence threshold is employed, the model might mistakenly eliminate parts that are, in fact, correct predictions. If not specified, the confidence threshold is set to 0.5 as the default setting.
Ablation on different modules The quantitative findings of this section are presented in Table 3. Initially, we fine-tuned SAM using ground truth bounding boxes directly obtained from the ground truth masks, representing the upper limit of our method. This approach yielded outstanding results, with an average Dice score surpassing . However, when we employed the most straightforward method of using the full image size as the bounding box input, SAM’s performance noticeably decreased to approximately . This decline highlights the significant influence of bounding box accuracy and quality on the overall segmentation performance of SAM.
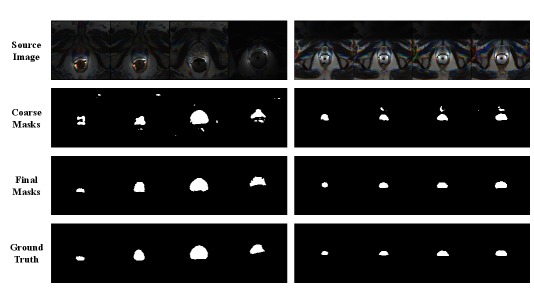
To address the need for precise bounding boxes, we applied the segmentation backbone network in section 3.2 to generate coarse masks and corresponding bounding boxes to fine-tune SAM. However, when we adopted the bounding boxes extracted from the coarse prediction masks directly, we did not observe any improvement in the results. Upon visual examination, we noticed a substantial presence of noise in the prediction masks (e.g. second row of Figure 2), which most likely contributed to the inaccurate bounding boxes. Therefore, we further employ the mask-filtering module described in section 3.3 to filter out the noise components from each mask, enabling the generation of refined bounding boxes. Fine-tuning SAM using these refined bounding boxes resulted in a significant enhancement of segmentation performance, reaching a Dice score of .
| Method | A to Rest | B to Rest | C to Rest | D to Rest | E to Rest | F to Rest | AVG |
|---|---|---|---|---|---|---|---|
| SAM+GT | 91.67 | 92.74 | 90.58 | 90.76 | 92.50 | 90.95 | 91.53 |
| SAM+Full | 77.82 | 78.19 | 67.46 | 71.04 | 78.02 | 73.94 | 74.41 |
| Resnet | 79.53 | 73.02 | 68.01 | 76.04 | 71.55 | 65.41 | 72.26 |
| SAM+Resnet | 81.76 | 75.44 | 63.65 | 77.14 | 63.81 | 71.98 | 72.30 |
| Final Result | 84.08 | 77.29 | 73.98 | 82.40 | 80.47 | 79.04 | 79.54 |
4.4 Qualitative Results
Figure 2 shows the qualitative outcomes of fine-tuning SAM using our approach on Domain E and Domain F of the Prostate dataset. We randomly selected four instances from each domain and applied SAM, fine-tuned on Domain F, to segment instances in Domain E, and vice versa. The results demonstrate that the process of predicting coarse masks and applying our mask-filtering module to generate refined bounding boxes can effectively enhance the accuracy of SAM during the fine-tuning process.
5 Conclusion
In conclusion, we have presented a novel framework that integrates SDG and SAM with mask-filtering postprocessing and merging strategy. We demonstrated the effectiveness of all the proposed modules and evaluated our approach to the Prostate dataset. The experiment showed that our method had greatly exceeded the state-of-the-art SDG methods in all directions of generalization on the dataset we trained. For future improvement, we plan to investigate more strategies to further reduce calculation costs and apply the model to other segmentation tasks while enhancing the network’s performance.
References
- Chen et al. (2020) Chen Chen, Chen Qin, Huaqi Qiu, Cheng Ouyang, Shuo Wang, Liang Chen, Giacomo Tarroni, Wenjia Bai, and Daniel Rueckert. Realistic adversarial data augmentation for mr image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part I 23, pp. 667–677. Springer, 2020.
- DeVries & Taylor (2017) Terrance DeVries and Graham W Taylor. Improved regularization of convolutional neural networks with cutout. arXiv preprint arXiv:1708.04552, 2017.
- Hu et al. (2023) Mingzhe Hu, Yuheng Li, and Xiaofeng Yang. Skinsam: Empowering skin cancer segmentation with segment anything model. arXiv preprint arXiv:2304.13973, 2023.
- Hu et al. (2022) Shishuai Hu, Zehui Liao, Jianpeng Zhang, and Yong Xia. Domain and content adaptive convolution based multi-source domain generalization for medical image segmentation. IEEE Transactions on Medical Imaging, 42(1):233–244, 2022.
- Huang et al. (2020) Zeyi Huang, Haohan Wang, Eric P Xing, and Dong Huang. Self-challenging improves cross-domain generalization. In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part II 16, pp. 124–140. Springer, 2020.
- Kirillov et al. (2023) Alexander Kirillov, Eric Mintun, Nikhila Ravi, Hanzi Mao, Chloe Rolland, Laura Gustafson, Tete Xiao, Spencer Whitehead, Alexander C Berg, Wan-Yen Lo, et al. Segment anything. arXiv preprint arXiv:2304.02643, 2023.
- Li et al. (2023) Chengyin Li, Prashant Khanduri, Yao Qiang, Rafi Ibn Sultan, Indrin Chetty, and Dongxiao Zhu. Auto-prompting sam for mobile friendly 3d medical image segmentation. arXiv preprint arXiv:2308.14936, 2023.
- Lin et al. (2023) Xian Lin, Yangyang Xiang, Li Zhang, Xin Yang, Zengqiang Yan, and Li Yu. Samus: Adapting segment anything model for clinically-friendly and generalizable ultrasound image segmentation. arXiv preprint arXiv:2309.06824, 2023.
- Liu et al. (2021) Quande Liu, Cheng Chen, Jing Qin, Qi Dou, and Pheng-Ann Heng. Feddg: Federated domain generalization on medical image segmentation via episodic learning in continuous frequency space. In IEEE Conference on Computer Vision and Pattern Recognition, pp. 1013–1023, 2021.
- Ma & Wang (2023) Jun Ma and Bo Wang. Segment anything in medical images. arXiv preprint arXiv:2304.12306, 2023.
- Ouyang et al. (2022) Cheng Ouyang, Chen Chen, Surui Li, Zeju Li, Chen Qin, Wenjia Bai, and Daniel Rueckert. Causality-inspired single-source domain generalization for medical image segmentation. IEEE Transactions on Medical Imaging, 42(4):1095–1106, 2022.
- Shaharabany et al. (2023) Tal Shaharabany, Aviad Dahan, Raja Giryes, and Lior Wolf. Autosam: Adapting sam to medical images by overloading the prompt encoder. arXiv preprint arXiv:2306.06370, 2023.
- Su et al. (2023) Zixian Su, Kai Yao, Xi Yang, Kaizhu Huang, Qiufeng Wang, and Jie Sun. Rethinking data augmentation for single-source domain generalization in medical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 37, pp. 2366–2374, 2023.
- Wang et al. (2020) Shujun Wang, Lequan Yu, Kang Li, Xin Yang, Chi-Wing Fu, and Pheng-Ann Heng. Dofe: Domain-oriented feature embedding for generalizable fundus image segmentation on unseen datasets. IEEE Transactions on Medical Imaging, 39(12):4237–4248, 2020.
- Wu et al. (2023) Junde Wu, Rao Fu, Huihui Fang, Yuanpei Liu, Zhaowei Wang, Yanwu Xu, Yueming Jin, and Tal Arbel. Medical sam adapter: Adapting segment anything model for medical image segmentation. arXiv preprint arXiv:2304.12620, 2023.
- Xu et al. (2022) Yanwu Xu, Shaoan Xie, Maxwell Reynolds, Matthew Ragoza, Mingming Gong, and Kayhan Batmanghelich. Adversarial consistency for single domain generalization in medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 671–681. Springer, 2022.
- Xu et al. (2020) Zhenlin Xu, Deyi Liu, Junlin Yang, Colin Raffel, and Marc Niethammer. Robust and generalizable visual representation learning via random convolutions. arXiv preprint arXiv:2007.13003, 2020.
- Zhang & Jiao (2023) Yichi Zhang and Rushi Jiao. How segment anything model (sam) boost medical image segmentation? arXiv preprint arXiv:2305.03678, 2023.
- Zhou et al. (2021) Kaiyang Zhou, Yongxin Yang, Yu Qiao, and Tao Xiang. Domain generalization with mixstyle. arXiv preprint arXiv:2104.02008, 2021.
- Zhou et al. (2022) Ziqi Zhou, Lei Qi, and Yinghuan Shi. Generalizable medical image segmentation via random amplitude mixup and domain-specific image restoration. In European Conference on Computer Vision, pp. 420–436. Springer, 2022.