Magneto: Combining Small and Large Language Models for Schema Matching
Abstract.
Recent advances in language models (LMs) open new opportunities for schema matching (SM). Recent approaches have shown their potential and key limitations: while small LMs (SLMs) require costly, difficult-to-obtain training data, large LMs (LLMs) demand significant computational resources and face context window constraints. We present Magneto, a cost-effective and accurate solution for SM that combines the advantages of SLMs and LLMs to address their limitations. By structuring the SM pipeline in two phases, retrieval and reranking, Magneto can use computationally efficient SLM-based strategies to derive candidate matches which can then be reranked by LLMs, thus making it possible to reduce runtime while improving matching accuracy. We propose (1) a self-supervised approach to fine-tune SLMs which uses LLMs to generate syntactically diverse training data, and (2) prompting strategies that are effective for reranking. We also introduce a new benchmark, developed in collaboration with domain experts, which includes real biomedical datasets and presents new challenges for SM methods. Through a detailed experimental evaluation, using both our new and existing benchmarks, we show that Magneto is scalable and attains high accuracy for datasets from different domains.
PVLDB Reference Format:
PVLDB, 18(8): 2681 - 2694, 2025.
doi:10.14778/3742728.3742757
††This work is licensed under the Creative Commons BY-NC-ND 4.0 International License. Visit https://creativecommons.org/licenses/by-nc-nd/4.0/ to view a copy of this license. For any use beyond those covered by this license, obtain permission by emailing info@vldb.org. Copyright is held by the owner/author(s). Publication rights licensed to the VLDB Endowment.
Proceedings of the VLDB Endowment, Vol. 18, No. 8 ISSN 2150-8097.
doi:10.14778/3742728.3742757
PVLDB Artifact Availability:
The source code, data, and/or other artifacts have been made available at %leave␣empty␣if␣no␣availability␣url␣should␣be␣sethttps://github.com/VIDA-NYU/magneto-matcher.
1. Introduction
The rapid increase in the volume of structured data– from data published in scientific articles (Nature, 2024; Science, 2024) and repositories (National Cancer Institute, 2024a; pdc, 2023; ukbio, 2024; European Organization For Nuclear Research and OpenAIRE, [n.d.]) to open government portals (of New York, 2024; datagov, 2024) – creates new opportunities to answer important questions through analytics and predictive modeling. But often, data from multiple sources must be integrated to answer these questions. Consider the following example.
Example 1.1.
In the context of the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) (cptac, 2024), Li et al. (2023) carried out a comprehensive proteogenomic analysis of cancer tumors. They collected data from ten studies (published as supplementary material in research papers) that cover multiple patient cohorts and cancer types. To facilitate their analysis, they mapped each dataset into the GDC standard, a data model set by the National Cancer Institute’s Genomic Data Commons (GDC) for cancer genomic data (Heath et al., 2021; National Cancer Institute, 2024a). Even though the studies had been carried out by members of the CPTAC effort, the datasets containing patient case and sample data used different representations for variable names and values. Integrating these data required a substantial effort to match variables from each dataset source schemata to the target GDC format, which encompasses over 700 attributes. ∎
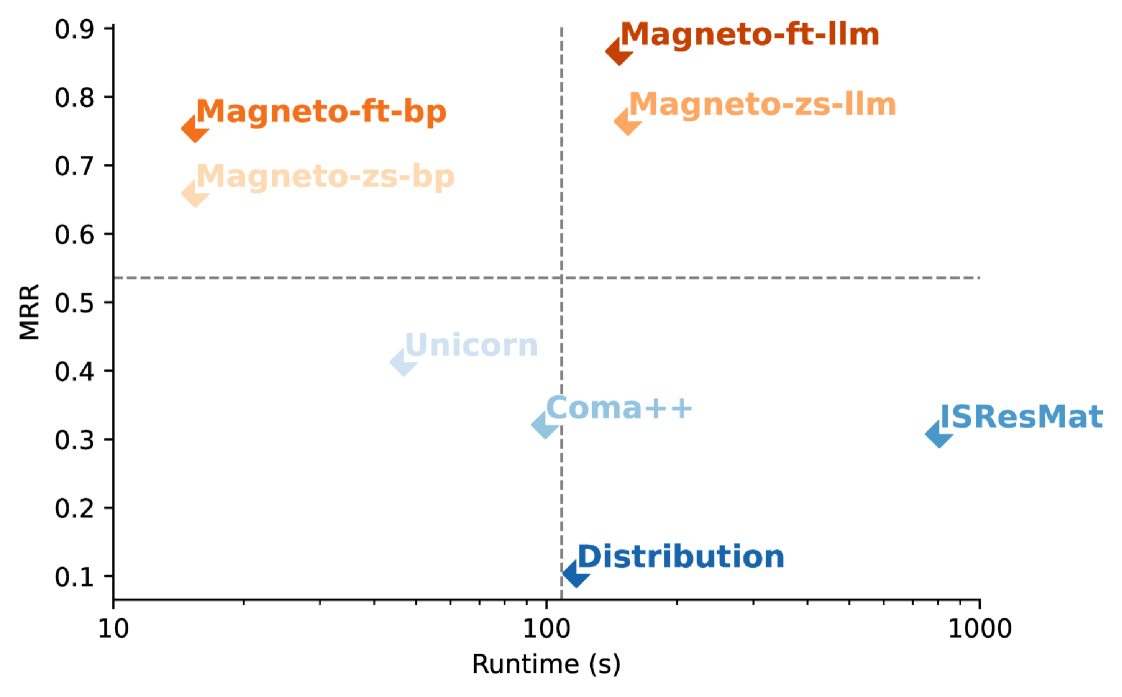
Even though the schema matching problem has been extensively studied (Doan et al., 2012; Cafarella et al., 2009; Miller, 2018; Koutras et al., 2021), matching still requires a time-consuming, manual curation process for complex tasks, which like the one described above, involves ambiguity and heterogeneity in the representation of attributes and values (see Table 1). Schema matching approaches that rely on attribute names, data types and values for similarity assessments are likely to fail for such matches. As a point of reference, we assessed the effectiveness of state-of-the-art strategies for the biomedical datasets from Example 1.1. As shown in Figure 1, they perform poorly: the best technique achieves at most 0.45 of mean reciprocal rank (MRR) and incurs high computational costs. We discuss these results in detail in Section 6.
Determining correspondences between columns may require knowledge beyond the schema and contents of a table. Table 1 shows some possible matches for the proteogenomic analysis. Sometimes, selecting the correct match is difficult even for subject matter experts. For example, for the attribute patient_age from one of the datasets, there are at least three plausible matches in GDC: age_at_diagnosis, days_to_birth, and age_at_index. Without additional context, it is difficult to determine the correct match.
Schema Matching and Language Models. Renewed interest in data integration has emerged due to the capabilities of language models (Feuer et al., 2024; Narayan et al., 2022). For schema matching, promising approaches have been proposed (Castro Fernandez et al., 2018; Cappuzzo et al., 2020; Du et al., 2024; Tu et al., 2023). A key challenge in schema matching is estimating the similarity between two columns. Pre-trained Language Models (PLMs), referred to as SLMs in this paper to distinguish them from large language models (LLMs), create column representations (or embeddings) enriched with semantic information. The similarity between two embeddings serve as a proxy for column-matching scores (Castro Fernandez et al., 2018; Cappuzzo et al., 2020). SLMs have also been fine-tuned for schema matching (Du et al., 2024) and general matching tasks (Tu et al., 2023).
| Source Column and Values | Target Candidate Columns |
| Histologic_Grade_FIGO: FIGO grade 1, FIGO grade 2, FIGO grade 3 |
tumor_grade: G1, G2, G3
who_nte_grade: G1, G2, G3 adverse_event_grade: Grade 1, Grade 2, Grade 3 |
| Path_Stage_Primary_Tumor-pT: pT1a (FIGO IA), pT2 (FIGO II), pT3b (FIGO IIIB) |
uicc_pathologic_t: T1a, T2, T3b
ajcc_pathologic_t: T1a, T2, T3b |
| Age: 65, 72, 49 |
age_at_diagnosis: n/a
days_to_birth: n/a age_at_index: n/a |
LLMs, in contrast, are trained using large, generic data corpora and thus contain knowledge that can assist in obtaining additional semantics necessary to identify matches. Prompting strategies combined with fine-tuned models have been shown effective to improve table understanding and help in integration tasks (Li et al., 2024a).
While prior work has shown the usefulness of SLMs and LLMs for schema matching, they also present significant practical challenges:
-
Challenge 1: Fine-tuning SLMs can lead to significant performance improvements, but this requires the availability of manually curated training data, which can be expensive to create.
-
Challenge 2: LLMs usually do not need fine-tuning, but face constraints due to fixed context windows, requiring truncation of large prompts and potential loss crucial information. While some models offer larger windows, cost scales with input and output size. Furthermore, accuracy can decline with long prompts and API calls incur high latency, especially for large inputs (Li et al., 2024b).
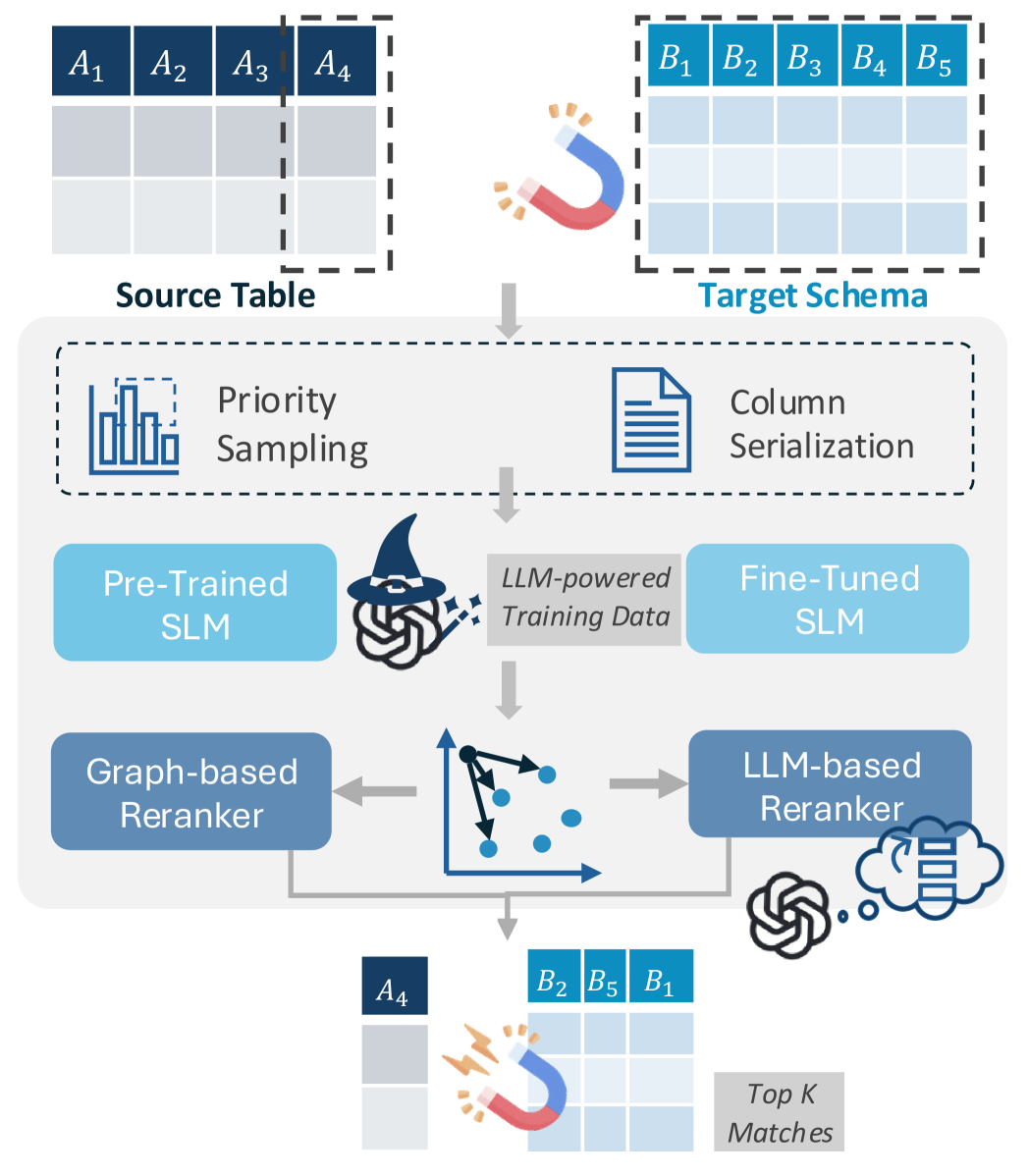
Our Approach. We introduce Magneto, a framework that combines SLMs and LLMs to derive cost-effective and general schema matching solutions. As illustrated in Figure 2, Magneto is structured in two phases: candidate retrieval selects a subset of the possible matches; and reranker, which ranks the candidates to make it easier for users to examine and select matches. To address Challenge 1, Magneto leverages LLMs to automate SLMs fine-tuning. Instead of relying on manually created training data and structure-based augmentation (e.g., row shuffling and sampling (Fan et al., 2023; Du et al., 2024)), we use LLMs to derive data. Using LLMs, we can add syntactic diversity and capture different representations for column names and their values. Magneto addresses Challenge 2 by using cheaper SLM-based methods for finding candidates thereby reducing the number of matches that need to be checked by more costly LLM-based rerankers. To ensure that all necessary details are included in the prompt while staying within the context window limit, we need to create representative samples of the columns in candidate matches to be ranked by the LLM. In addition, since LLMs are designed for textual data, we must serialize the column content. Selecting the right serialization strategy is still an open research problem that has attracted substantial attention (Fang et al., 2024; Dong and Wang, 2024). We explore different alternatives for both sampling and serialization.
Our goal with Magneto is to achieve high accuracy for schema matching at a low cost. Figure 1 illustrates the trade-offs between runtime and accuracy for existing schema matching algorithms and different variants of Magneto. Note that traditional methods tend to cluster at relatively high accuracy with long runtimes or relatively low runtimes with reduced accuracy. In contrast, Magneto variants strike a balance across both dimensions and have the advantage of requiring no training data.
We conduct an extensive experimental study of Magneto on various datasets from two benchmarks. The first is a new benchmark, that we developed in collaboration with domain experts, which includes real biomedical datasets. Our experiments show that many existing schema matching solutions struggle with this benchmark, particularly given the complexity and syntactic variability present in its datasets. In contrast, Magneto demonstrates superior accuracy and generally faster runtimes. We also evaluate Magneto on the Valentine benchmark (Koutras et al., 2021) and observe that it also performs well for the Valentine datasets which cover a variety of domains.
Contributions. Our main contributions are summarized as follows:
-
We introduce the Magneto framework that effectively combines SLMs and LLMs and allows the creation of schema matching strategies that attain a balance between accuracy and runtime for diverse data and tasks (Section 2).
-
We propose an LLM-powered method to generate training data for fine-tuning SLMs for schema matching tasks and a contrastive learning pipeline using triplet loss and online triplet mining to enhance column embedding distinction (Section 3).
-
Unlike previous approaches that use LLMs for schema matching, we show that reranking candidate matches with LLMs, derived from less costly methods, achieves high accuracy at a lower cost. (Section 4).
-
We perform an extensive experimental evaluation of different strategies derived using our framework, comparing them against SM methods over datasets that cover a wide range of domains. We also carry out ablation studies to assess the effectiveness of different choices for column sampling and serialization (Section 6).
2. Motivation and Solution Overview
Biomedical Data Integration: Opportunities and Challenges. Due to substantial investments made in infrastructure to share biomedical data (National Institutes of Health, 2020), many datasets are now stored in repositories with related research articles (pdc, 2023; National Cancer Institute, 2024a). Pooling these data can advance research across various diseases and populations, but current technologies are limited in their ability to integrate these data. Our research was motivated by this challenge as part of a collaboration with biomedical researchers in the context of the ARPA-H Biomedical Data Fabric (BDF) program (ARPA-H, 2024), which aims to develop usable data integration methods and tools.
Our interviews with biomedical experts revealed that current data integration practices rely heavily on manual processes and dataset-specific scripts for schema matching (Mertins et al., 2016; Li et al., 2023; Cheng et al., 2024). As illustrated in Example 1.1, this approach is both error-prone and difficult to reproduce, creating significant barriers to biomedical research.
Based on these findings, we identified two critical requirements that guided the design of Magneto. First, researchers need approaches that generalize across the diverse landscape of biomedical data—spanning different data types (genomics, proteomics, electronic health records), disease categories (various cancers, autoimmune and rare diseases), and data sources (publications, hospitals, data commons). Second, given the complexity of biomedical schemas, these approaches must facilitate curation, as even subject matter experts often struggle to determine correct matches.
2.1. Definitions and Evaluation Metrics
Before describing our approach, we introduce the notation, schema matching definition, and evaluation metrics used in this paper.
Definition 2.0.
(Schema Matching) Let be a source table and be a target table, where and are columns that define the schemata. Each column has an associated domain, denoted , representing the set of possible values that the column can take; note that may be empty. Schema matching focuses on aligning the table schemata by establishing correspondences between columns representing the same real-world concept or entity. A matching algorithm (or matcher) aims to identify pairs that represent the same (or semantically equivalent) column based on various factors, such as their domains and names. Thus, a matcher can be seen as a function that generates a schema mapping , where each element represents a correspondence between a source column and a target column , where , meaning that the domains of columns and are related or overlap. ∎
Matching algorithms often associate a score with each match they derive. These scores are used to generate ranked lists containing the derived matches, which help users explore matches by prioritizing the highest-scoring candidates. These lists provide a global ranking of best matches among all pairs of possible matches of a given pair of tables S and T or a per-column ranking of the best matches for a given source attribute . Thus, a common approach to evaluate schema matching methods is to assess their ability to produce high-quality ranked lists of matches (Koutras et al., 2021; Du et al., 2024). We use two evaluation metrics: Mean Reciprocal Rank (MRR) and Recall at Ground-Truth Size (Recall@GT), as detailed next.
Definition 2.0.
(Mean Reciprocal Rank) Let denote the ranked list of matches produced by a schema-matching algorithm for the source column , and be the position of the first correct target column within the ordered list . The reciprocal rank (RR) of an individual column is the multiplicative inverse of the rank, i.e., . The mean reciprocal rank (MRR) for a table S is the average RR over the subset of columns that contain a correct match in the ground truth:
| (1) |
Intuitively, MRR measures how long it takes to find the first relevant match when examining the ranked list of matches. ∎
MRR is a standard evaluation metric for ranked lists in search engines and question-answering systems (Schütze et al., 2008; Voorhees et al., 1999). For schema matching, high-quality results correspond to ranking the most relevant match for each source column as highly as possible. A high MRR score therefore indicates that users can more easily identify correct matches when evaluating candidate matches for a given column.
We also use the recall at ground truth size (Recall@GT), a standard measure used in recent schema matching literature (Koutras et al., 2021). Unlike MRR, which evaluates rankings per source column, Recall@GT operates on a global ranking that merges all candidate matches across columns into a single list.
Definition 2.0.
(Recall@GT) Let denote the global ranked list containing all matches produced by a matcher that considers all pairs of possible matches between columns from S and T, and let denote a set containing only the top- best results in . Moreover, let be the set of ground truth matching pairs . Recall@GT measures the fraction of relevant matches in the ground truth that also appears in , where is given by the size of the ground truth, . More formally,
| (2) |
Intuitively, it measures how well the matcher can place all correct matches of a table S at the top of the global ranked list. ∎
2.2. Magneto: Overview
Magneto first applies a cheaper approach to retrieve and filter candidates so that a more sophisticated method can accurately identify the correct matches from a smaller candidate set. Figure 2 shows one variant of Magneto that uses a fine-tuned language model as the retriever and a large language model as the reranker.
Candidate Retriever. The candidate retriever leverages an SLM to generate a ranked list of potential matches from the target table for each input column. It uses column embeddings to estimate column pair similarities (Cappuzzo et al., 2020; Fan et al., 2023). SLMs are a good choice for this step given their ability to capture semantic similarity and efficiency. General-purpose pre-trained SLMs such as BERT (Devlin, 2018), RoBERTa (Liu, 2019), and MPNet (Song et al., 2020), may struggle with syntactic differences and lack contextual knowledge for domains absent in their training data. To perform complex tasks effectively, these models need to be fine-tuned. We propose to leverage LLMs to automatically generate the data needed to fine tune SLMs. We show that LLMs can help derive high-quality training data that reflect instances of variability for semantically similar columns that arise in real data. This approach, described in Section 3, improves the robustness of the SLM-based retriever, making it possible for it to handle complex matches without requiring human-labeled training data. We refer to Magneto configurations that use this approach using the label ft. Note that Magneto-ft invokes LLMs only during the offline training phase for a given domain. Once trained, the fine-tuned SLM can perform multiple inferences efficiently without making calls to LLMs.
Match Reranker. While fine-tuning improves SLM performance, it is not sufficient to handle schema matching tasks involving domains and heterogeneity unseen during the fine tuning. To improve generalizability, Magneto uses LLMs as rerankers to refine the candidates identified by the SLM-based retriever. This approach, which we refer to as Magneto-llm, enhances accuracy by leveraging carefully designed prompts and techniques that enable the LLM to judiciously assess matches and discern subtle semantic nuances, which are challenging for the SLM to detect independently. As discussed in Section 4, Magneto-llm also reduces LLM costs.
Varying Retrievers and Rerankers. The architecture of Magneto allows the combination of different alternatives for retrievers and rerankers. We can combine traditional matching techniques with the embedding strategies for the candidate retriever. For example, when columns in a match have the same name, modulo minor variations in case and punctuation, we assign a perfect similarity score of 1.0. During our experiments, this simple technique consistently improved overall accuracy, albeit slightly, in most cases.
We also implemented bipartite-graph reranker as an algorithmic alternative that adapts the filtering technique from Melnik et al. (Melnik et al., 2002). This approach, which we refer as Magneto-bp, is particularly suitable for scenarios where LLMs are unavailable during inference or where strict runtime constraints must be met. The algorithm combines all ranked match candidates across source columns into a single global list and transforms it into an undirected bipartite graph. There are two (disjoint) node sets consisting of the columns in the source and target tables (respectively). The graph contains two disjoint node sets representing the source and target table columns, respectively, with potential matches represented as weighted edges between nodes. Edge weights correspond to match confidence scores. To identify the best match for the set of attributes in the source column, it uses the algorithm from (Crouse, 2016), which solves the assignment problem in polynomial time and scales to large graphs. The final ranking prioritizes matches selected by the assignment algorithm at the top of the list, while unselected matches are placed at the bottom in their original relative order.
3. Using LLMs to Fine-Tune SLMs
Pre-trained small language models (SLMs) have been used to help with schema-matching related tasks by encoding semantic information from column names and values into dense vector representations–embeddings (Tu et al., 2023; Zhang et al., 2023; Fan et al., 2023; Cong et al., 2023; Dong et al., 2023; Du et al., 2024). To identify matches, embeddings of source and target columns can be compared using cosine similarity: high similarity scores indicate a higher likelihood that the columns match. SLMs work well for general natural language tasks, but their ability to interpret tabular data is limited. These models must often be fine-tuned to handle tasks that involve tables. However, fine-tuning approaches require large amounts of labeled data for training (Tu et al., 2023; Dong et al., 2023). Unfortunately, this is impractical in many scenarios such as the integration of biomedical data (Example 1.1), for which training data is hard to obtain.
In the absence of training data, it is possible to apply augmentation techniques to automatically generate variations of data to be used as positive examples. For example, given a column, different versions can be generated by shuffling rows, sampling values, and applying perturbations to values (Fan et al., 2023; Dong et al., 2023). However, these variations may not fully capture the heterogeneity found in real data. We introduce a new method that leverages LLMs to generate training data and present a pipeline to fine-tune SLMs for schema matching (Section 3.2). Another key consideration is how to represent tables and their columns, which we discuss in Section 3.1.
3.1. Value Sampling and Column Serialization
Priority Sampling for Column Values. We generate embeddings to retrieve candidate matches by including a sample of column values in the column representation. Sampling strategies have been adopted for data management tasks, including column type annotation (Feuer et al., 2024) and table union search (Fan et al., 2023). A common approach is to use weighted sampling and assign higher weights to more frequent values. We adopt this approach and incorporate coordination into the sampling process with priority sampling (Daliri et al., 2024). For inner product sketching, priority sampling maximizes the likelihood of selecting corresponding values across vectors by emphasizing elements with larger magnitudes.
In our setting, priority sampling is adapted to optimize the selection of column values. This approach not only prioritizes frequent values, which are statistically more representative of the column domain, but it also increases the likelihood of identifying shared values across different columns that act as inter-column anchors. These anchors enhance similarity detection in SLMs, which are sensitive to token co-occurrence patterns learned during pre-training. Specifically, we use a random seed to select a uniformly random hash function , where represents the maximum number of unique values across all columns. For each value , we compute a rank:
where denotes the frequency of . Then, we select the sample’s first values with the largest priorities .
Column Serialization. Since SLMs interpret inputs as sequences of tokens (i.e., regular text), we must transform each column into a token sequence that the model can process. Recent research has explored various approaches to serializing columns into a textual format (Badaro et al., 2023). Here, we explore the impact of different approaches to column serialization for schema matching.
Given column with column type and sample values , we consider the following serialization approaches:
where the [CLS] token is a special symbol that indicates the start of a column, and [SEP] separates column components.
is a variant of a widely-adopted serialization strategy (Suhara et al., 2022; Yin et al., 2020). Instead of employing commas to separate sample values, we utilize the [SEP] token. This modification prevents the model from interpreting the values as an ordered text sequence, thereby treating them as unordered, discrete features. This change is crucial in schema matching as it emphasizes the structured and independent nature of the data in each column.
is an extension of where prefixes are added to delineate each component and provide additional context for the SLM. By explicitly tagging each data segment, it helps models better contextualize the information, leading to improved interpretability and alignment accuracy in schema matching tasks.
repeats the column name multiple times to reinforce its importance, nudging the model to prioritize column names. This strategy is inspired by findings in attention-based neural network research where repetition can enhance item salience (Vaswani et al., 2017). It attempts to mitigate attention drift in SLMs, which can disproportionately focus on later tokens or values rather than column names, especially in zero-shot settings where the model is not optimized to learn the importance of different column components. For our experiments, we set to strike a balance between reinforcing column names without overwhelming the model with redundancy.
In addition to column name and values, we incorporate data types into the column representation. For column type inference, we classify columns into basic types: numerical, categorical, date, or binary. We classify columns with a high proportion of unique values (e.g., over 90% distinct values) as “key” columns, as they often represent unique identifiers. When column value is unavailable or type detection fails, we label the column type as “unknown”.
In Magneto, we use serialization as a hyperparameter. We study the impact of different serialization strategies for schema matching in Section 6, where we also investigate different approaches to sampling column values. We experimented with varying column value sample sizes (from 10 to 30) and observed only marginal differences. Interestingly, smaller sample sizes (e.g., ten values) sometimes outperformed larger ones, likely because of reduced noise. Thus, we fixed the sample size to 10 as a default.
3.2. LLM-Powered Fine Tuning
By fine-tuning, we aim to learn column representations whose embeddings cluster spatially to reflect semantic relationships. This spatial arrangement enables efficient retrieval of related columns via cosine similarity. Our training methodology thus minimizes the distance between embeddings of identical or semantically related columns while maximizing separation between distance ones.
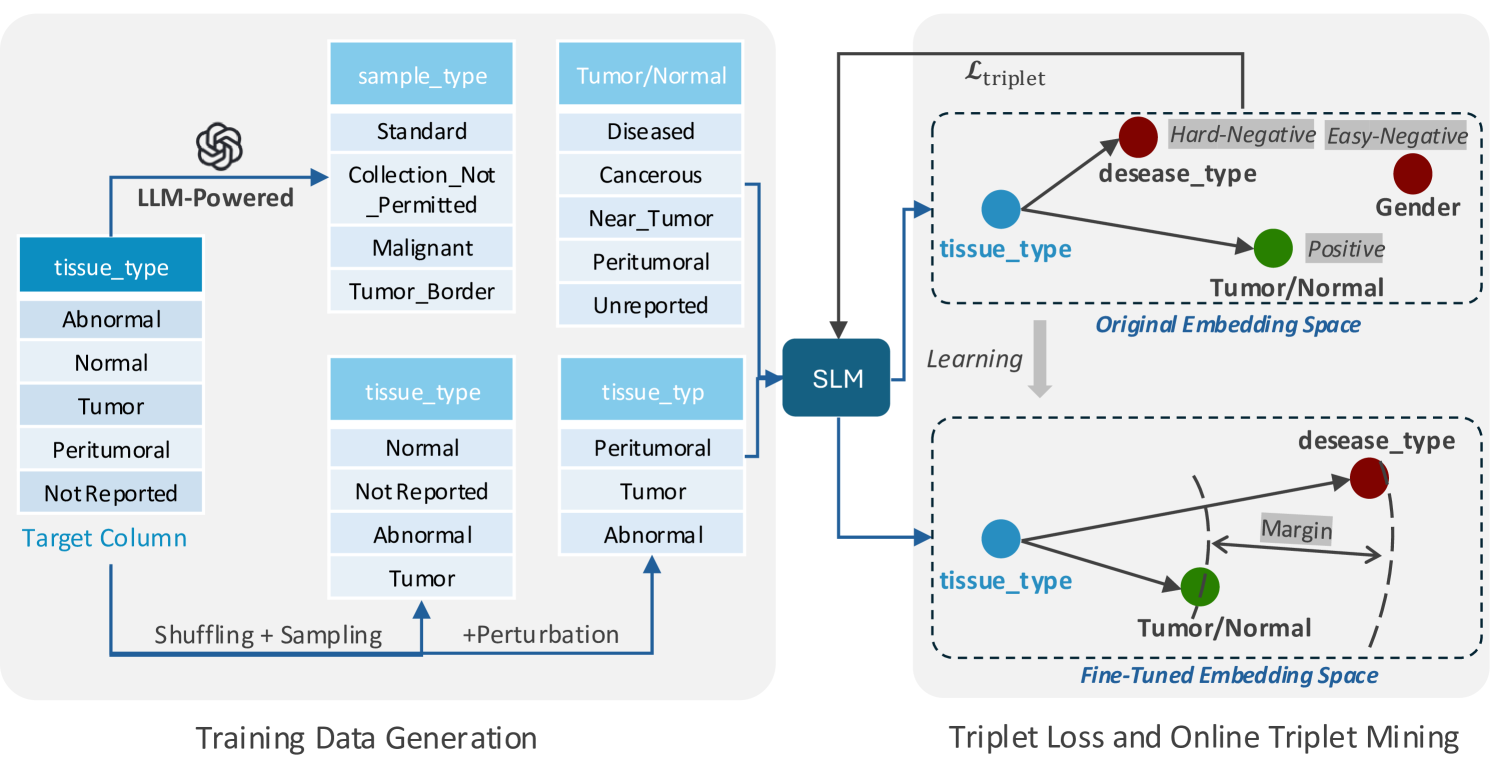
LLM-Derived Training Data. As discussed above, an important challenge in fine-tuning lies in obtaining high-quality training data. With the goal of capturing syntactic heterogeneity common in real datasets, we introduce LLM-based augmentation (llm-aug), an approach that generates variations of columns that are semantically equivalent but syntactically different. The derived columns serve as the training data fine-tuning process, which adopts a self-supervised contrastive learning approach (Figure 3).
Synthetic columns are derived from an input (anchor) column as follows: 1) The anchor name and a sample of its values are given to the LLM using the structured prompt depicted in Figure 4, and 2) the LLM outputs columns that are semantically similar to the anchor. Examples of synthetic columns are shown in Table 2. This method establishes a class of columns considered matches (derived from the same anchor); columns within the same class serve as positive examples and those from different classes as negative examples.
| Instruction | Given the table column [Column Name] with values [Sample Column Values], generate three alternative column names that adhere to typical database naming conventions such as underscores and abbreviations. Additionally, provide distinct, technically correct synonyms, variants, or abbreviations for the listed values. For columns with numerical or datetime data, generate random numbers or dates appropriate to the column’s semantic meaning. |
| Format | Ensure that each set does not exceed 15 values. Format your output as follows: alternative_name_1, value1, value2, value3, …; alternative_name_2, value1, value2, value3, …; alternative_name_3, value1, value2, value3, … Ensure your response excludes additional information and quotations. |
| Original Data | Generated Data | |
| Column: | tissue_source_sites | tumor_site |
| Values: | Thyroid, Ovary | Thyroidal, Ovarian |
| Column: | exon | gene_segments |
| Values: | exon11, exon15 | segment11, segment15 |
| Column: | masked_somatic_mutations | genetic_variants |
| Values: | MET_D1010N, FLT3_ITD | D1010N_MET, ITD_FLT3 |
| Column: | max_tumor_bulk_site | primary_tumor_location |
| Values: | Maxilla, Splenic lymph nodes | Maxillary, Splenic_nodes |
We combine the LLM-based augmentation with other structure-based augmentation methods (struct-aug) such as random sampling and shuffling of values, and minor perturbations to the column name, including random character replacements or deletions (Fan et al., 2023). These techniques inject variability while preserving syntactic and structural similarity to the original column. By using both augmentation strategies, the model can learn to identify matches with different characteristics. We restrict this fine-tuning and training process to the target table columns to avoid inaccuracies. Applying this process to source tables with unknown true matches to target tables could mistakenly classify them as negative examples, introducing errors into the embedding space.
Triplet Loss and Online Triplet Mining. To leverage the generated synthetic columns, we implement a contrastive learning framework using triplet loss and online triplet mining (Schroff et al., 2015). Unlike standard contrastive losses, triplet loss directly optimizes relative similarities, improving schema alignment precision.
Each training triplet includes an anchor column , a same-class positive , and a different-class negative . The model learns to minimize and maximize , where is cosine distance. Specifically, we incorporate Batch Hard triplet loss (Schroff et al., 2015) which is defined as follows:
| (3) |
Here, is a margin hyperparameter that ensures a minimum distance between the positive and negative embeddings relative to the anchor, thereby enhancing the separability between classes. The loss iterates over classes and columns within each training batch, optimizing the embeddings to emphasize inter-class distinctions and intra-class similarities.
Additionally, we apply Online Triplet Mining (Schroff et al., 2015; Hermans et al., 2017), which enhances the learning process by dynamically selecting the most challenging positive and negative examples within each training batch. This technique prioritizes triplets that maximize learning efficiency, specifically focusing on:
-
Hard Negatives: Closest negative to the anchor that violates the margin constraint:
-
Semi-Hard Negatives: Negatives farther than the positive but within the margin:
-
Hard Positives: Farthest positive from the anchor within its class:
These conditions help the model not settle for easy examples and instead learn to distinguish subtle differences, developing more robust and discriminative features to distinguish columns. Together, triplet loss and mining improve embedding discriminability for schema matching.
Model Selection for Schema Matching. To select the optimal fine-tuned model during training, we must define an effective validation metric specifically tailored for schema matching tasks. Our selection process uses the measures described in Section 2.1 to evaluate model performance on synthetic data. Since no ground-truth data is available, we rely on synthetic datasets to simulate real-world scenarios. These metrics are specifically chosen to ensure that the model identifies correct matches and ranks them in a manner that reflects their true relevance.
We compute a validation score that averages the MRR and recall at ground truth: We implement early stopping when the validation accuracy remains unchanged for 5 epochs, selecting the model with the highest validation score as the best fine-tuned model.
4. LLMs as Rerankers
SLMs can serve as efficient retrievers, but they may fail to capture complex semantic relationships. Moreover, models fine-tuned with LLM-derived data are inherently domain-specific. To mitigate these limitations, Magneto incorporates a Large Language Model (LLM) as a reranker. Unlike existing methods that adjust match rankings based on heuristic and similarity metrics (Madhavan et al., 2001; Melnik et al., 2002; Do and Rahm, 2002; Gal, 2006; Radwan et al., 2009; Alserafi et al., 2020), Magneto leverages LLM understanding to complement initial retrieval and improve overall schema matching performance.
A natural question arises: Why not just use an LLM for schema matching? While there are approaches that employ LLMs for schema matching (Li et al., 2024a; Parciak et al., 2024), their applicability in scenarios that involve many or large tables is limited due to high computational costs and challenges related to context windows (Challenge 2, Section 1). We also empirically demonstrate that LLM-only approaches underperform an SLM-LLM combination for large target schemas (Figure 11). We posit that LLMs must be used judiciously and designed Magneto accordingly.
The core of our reranking approach is a carefully designed prompt template that converts the abstract task of column-pair similarity assessment into a more structured and interpretable process. We designed this prompt inspired by recent lessons learned from column type annotation (Feuer et al., 2024) and trends in prompt engineering (White et al., 2023; Narayan et al., 2022). Figure 5 shows the structure of the prompt.
| Scoring-Oriented Instruction | From a score of 0.00 to 1.00, judge the similarity of the candidate column in the source table to each target column in the target table. Each column is represented by its name and a sample of its respective values, if available. |
|
One-shot
Example |
Example:
Candidate Column:: Column: EmpID, Sample values: [100, 101, 102] Target Schemas: Column: WorkerID, Sample values: [100, 101, 102] Column: EmpCode, Sample values: [00A, 00B, 00C] Column: StaffName, Sample values: [“Alice”, “Bob”, “Charlie”] Response: WorkerID(0.95); EmpCode(0.30); StaffName(0.05) |
| Format | Provide the name of each target column followed by its similarity score in parentheses, formatted to two decimals, and separated by semicolons. Rank the column-score pairs in descending order. Exclude additional information and quotations. |
| Input |
Candidate Column::[Serialized Source Column]
Target Schemas:[Serialized Target Columns] Response: |
Scoring-Oriented Prompt Design. The Scoring-Oriented Instruction aligns the model with schema matching by sending one source column and the top target candidates from the SLM to the LLM. Prior works (Li et al., 2024a; Parciak et al., 2024) use table-wise prompts that rank columns without individual scores, limiting scalability. Column-wise ranking without scoring also hinders table-wise comparisons, such as recall@GT evaluation. To address these limitations, we propose a novel prompt design that requires the model to assign scores from 0.00 to 1.00 to each column pair, rather than merely producing a ranked list. This facilitates direct comparisons across different pairs and allows the model to adopt a holistic view by considering all target columns simultaneously during scoring. Additionally, our serialization of the column data clarifies to the model that a text sequence comprising a column name followed by its values represents a column in a relational table. This allows more accurately scoring matches based not only on the semantic and contextual relevance but also on the structural characteristics of the data.
Few-Shot In-Context Learning. By providing a few examples of a task to an LLM, few-shot learning can lead to significant improvement in LLM performance (Brown et al., 2020; Dong et al., 2022). This method has been shown effective in various data management tasks, enabling robust performance without the need for extensive fine-tuning (Hegselmann et al., 2023; Shraga et al., 2020).
In the One-shot Example strategy, we provide a single example of the schema matching task, which outputs a ranked list with scores. This approach clarifies both the objective of the task and expected output format, and establishes a uniform scoring standard, enhancing the comparability of matches across all source and target column pairs. Utilizing only one example helps maintain a lightweight prompt structure, crucial for minimizing input and output token counts. The number of tokens directly impacts runtime and cost, and it can also influence accuracy (Li et al., 2024b).
Optimization of Model Cost. To ensure reliability, we attempt to parse the LLM’s output up to three times. If parsing fails after these attempts, we revert to using the embedding scores as a fallback mechanism. During testing, output-related issues were infrequent; however, they occurred more commonly with larger tables and in scenarios lacking a candidate retriever.
Note that the design of Magneto makes it possible to balance accuracy with computational cost. Based on operational constraints (or requirements), the number of candidates sent to the reranker can be adjusted. The scores for candidates not assessed by the reranker are normalized such that the maximum score aligns with the lowest score received from the reranker. Therefore, a complete ranked list can still be returned to the user.
5. The GDC benchmark
A major challenge in evaluating schema-matching algorithms is the lack of benchmarks reflecting the diversity of real-world data. Benchmarks are often synthetically fabricated from real data (Koutras et al., 2021; Das et al., [n.d.]), which can introduce biases. Moreover, as show in Section 6 (and in (Koutras et al., 2021)), publicly available benchmarks based on real data such as Magellan (Das et al., [n.d.]) and WikiData (Koutras et al., 2021) are quickly becoming “saturated” since many algorithms attain near-perfect performance and leave small room for algorithmic improvements. This makes it difficult to extract useful insights about the strengths of different algorithms.
To address this problem, we built a new benchmark dataset (Santos et al., 2025) based on the real data harmonization scenario described in Example 1.1. We collaborated with biomedical researchers to design a benchmark that reflects the challenges they face when working with biomedical data. We obtained datasets from ten studies related to tumor analysis (Cao et al., 2021; Clark et al., 2019; Dou et al., 2020; Gillette et al., 2020; McDermott et al., 2020; Huang et al., 2021; Krug et al., 2020; Satpathy et al., 2021; Vasaikar et al., 2019; Wang et al., 2021), and, with the experts’ help, manually aligned and matched these datasets to the Genomics Data Commons (GDC) standard (National Cancer Institute, 2024a).
The GDC is a program of the US National Institutes of Health responsible for handling genomic, clinical, and biospecimen data from cancer research initiatives. Its standard dictionary describes data using a graph model that includes names and descriptions for nodes and attributes and acceptable values for some attributes. To be compatible with existing schema matching solutions, we transformed the model to a relational schema that contains only column names and domain information (i.e., we disregard column descriptions). We created a simplified table reflecting the GDC format, the “target” table, listing domain values for each column without repetition.
Table 1 illustrates a few samples of the data published by Dou et al. (Dou et al., 2020) alongside their corresponding GDC format. As we can observe, matching clinical data with the GDC standard poses several challenges, including terminology mismatches and data format variations. The benchmark includes 10 pairs of source-target tables. The number of columns in the source tables ranges from 16 to 179, and the number of rows ranges from 93 to 225. Our simplified GDC target schema comprises a single table with 736 columns. While some columns have a small number of distinct values (e.g., binary yes/no attributes), some contain up to 4478 distinct values. The ground truth was manually curated by multiple annotators, who used a mix of manual and automated methods to identify possible candidate matches (e.g., GDC search tools (National Cancer Institute, 2024b) and bdi-kit (The bdi-kit contributors, 2024)). Given that the correctness of some matches is challenging to determine even for bioinformatics experts (e.g., it may require reading the original papers or asking data producers), the final match decisions were made by consensus based on what users would expect from an algorithm given the limited context.
6. Experimental Evaluation
6.1. Experimental Setup
Datasets. We evaluated Magneto on six datasets (Table 3), grouped as: (1) Human-Curated-reflecting real-world matches, and (2) Fabricated—systematically generated to capture structural variations and diverse match types. The GDC benchmark (Section 5) reflects challenges in biomedical data integration. The other five datasets are from the Valentine schema matching benchmark (Koutras et al., 2021) and have been used to evaluate recent schema matching solutions (Tu et al., 2023; Du et al., 2024).
| Dataset | #Table Pairs | #Cols | #Rows | Match Type |
| GDC | 10 | 16–736 | 93–4.5k | Human-Curated |
| Magellan | 7 | 3–7 | 0.9k–131k | Human-Curated |
| WikiData | 4 | 13–20 | 5.4k–10.8k | Human-Curated |
| Open Data | 180 | 26–51 | 11.6k–23k | Fabricated |
| ChEMBL | 180 | 12–23 | 7.5k–15k | Fabricated |
| TPC-DI | 180 | 11–22 | 7.5k–15k | Fabricated |
Baselines. We compare Magneto against several approaches, ranging from traditional string-similarity-based methods to sophisticated model-driven techniques. For traditional approaches, we use:
-
COMA: Combines multiple strategies to compute and aggregate the similarity of table metadata (Do and Rahm, 2002);
-
COMA++: An extension of COMA that leverages column values and their distributions (Aumueller et al., 2005);
-
Distribution: Detects column correspondences based on data value distribution (Zhang et al., 2011);
-
SimFlooding: Uses several graph-based techniques to identify correspondences between metadata (Melnik et al., 2002).
We evaluated the Jaccard-Levenshtein baseline proposed in (Koutras et al., 2021) and the Cupid algorithm (Madhavan et al., 2001). However, both methods consistently performed significantly worse than other approaches across all experiments, so we excluded them from the plots for clarity. We used the implementations available in the Valentine repository for all traditional approaches (Koutras et al., 2021).
We also compared Magneto against recent approaches that use LMs: ISResMat which leverages contrastive learning and embeddings from SLMs (Du et al., 2024), and Unicorn, a supervised and general approach using language models for data encoding and trained for data integration tasks (Tu et al., 2023). We used the implementation from the authors for ISResMat. For Unicorn, we used the pre-trained model and implementation provided by the authors. The match scores were derived from the model’s predicted match probabilities. It is important to note that Unicorn requires external training data for optimal performance. The zero-shot version performed significantly worse than other baselines and was excluded from our evaluation. We report Unicorn numbers for the algorithm in Valentine datasets for completeness, but note that its model was trained on the fabricated datasets from Valentine (Koutras et al., 2021), so data leakage can influence the reported results.
Implementation Details. The COMA algorithm is implemented in Java. Magneto and all other methods are written in Python. We use MPNet as our underlying small language model. It is pre-trained on masked and permuted language tasks, enabling precise contextual understanding of unordered text in column headers and entries (Song et al., 2020; Dong et al., 2023; Sheetrit et al., 2024). For the LLM reranker, we use the GPT-4o-mini model from the OpenAI API due to its robust performance and cost-effectiveness. However, these choices are flexible; alternative models can be easily integrated as replacements based on cost or performance requirements. It is important to note that our study does not focus on determining the optimal model for this task.
We assess four variations of Magneto that represent different combinations of retrievers and rerankers: Magneto-zs-bp (zero-shot SLM, bipartite reranker), Magneto-ft-bp (fine-tuned SLM, bipartite reranker), Magneto-zs-llm (zero-shot SLM, LLM reranker), Magneto-ft-llm (fine-tuned SLM, LLM retriever).
We set margin for the fine-tuning configuration, and run the model for 30 epochs on the GDC benchmark and 10 epochs on the Valentine datasets.
Execution platform. The experiments were run on a server running Red Hat Enterprise Linux version 9.2. We used a single node consisting of 8 cores and 64 GB of memory for CPU-based tasks. The GPU-intensive experiments used an NVIDIA A100 GPU. For Magneto variations, Unicorn, and IsResMat, we used a GPU to run all experiments, except the scalability experiment described in Section 6.3, conducted on a CPU for consistency with other methods.
6.2. End-to-End Accuracy
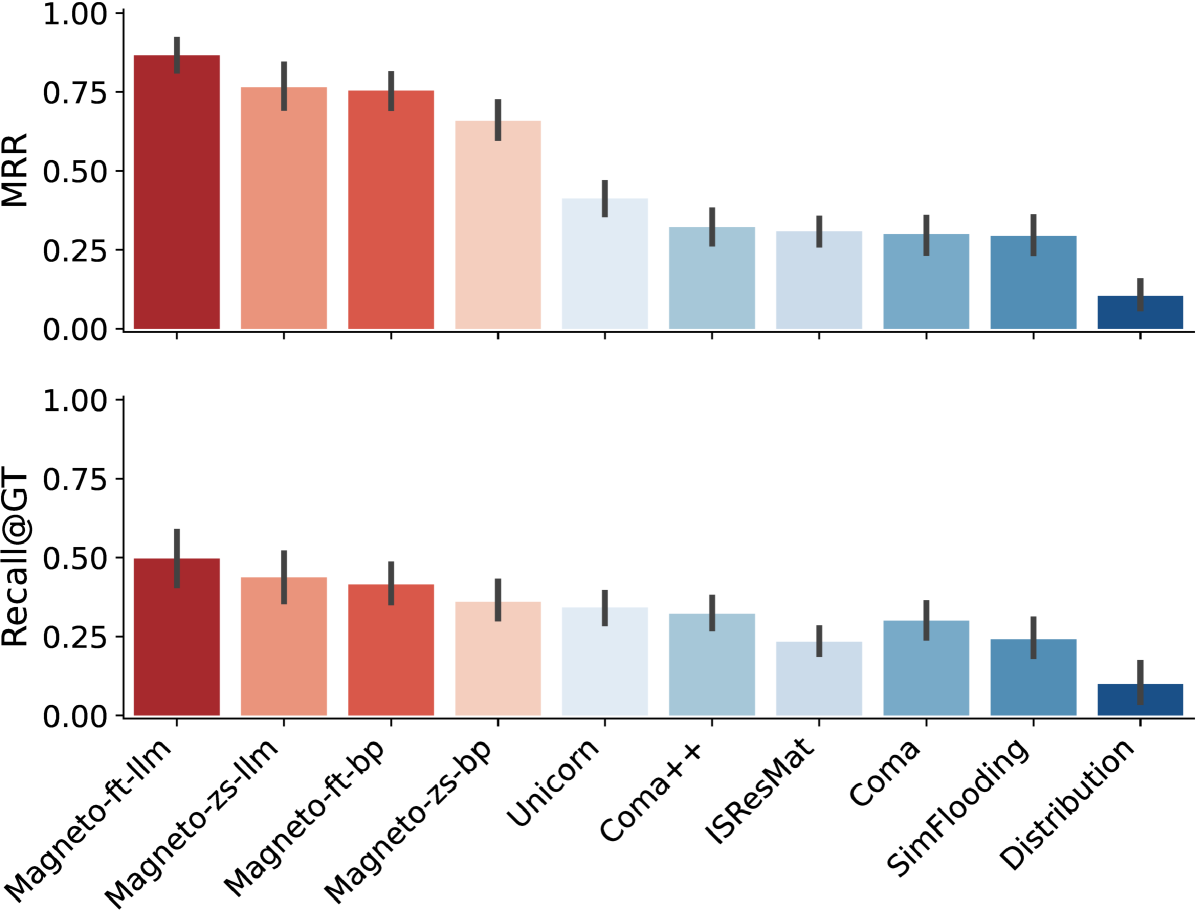
Performance on GDC. Figure 6 shows the accuracy (measured by MRR and Recall@GT) for all versions of Magneto and the baseline methods for the GDC benchmark. The Magneto variations outperform all other methods for both measures. Magneto-ft-llm has the highest overall accuracy, confirming that the combination of a fine-tuned SLM and an LLM is effective for this complex matching task. The fact that the zero-shot Magneto variations also outperform recent state-of-the-art approaches demonstrates the effectiveness of the serialization and sampling techniques we designed for schema matching. We ablate the different components of Magneto in Section 6.4, where we discuss this in more detail.
Performance on Valentine. The accuracy results for the Valentine benchmark are shown in Figure 7. Note that the different methods attain much higher accuracies for these datasets than for GDC. The Valentine datasets are derived either from open data sources or synthetic repositories, and they are also meticulously curated. In contrast, the GDC benchmark includes real scientific datasets and features a diverse set of column names and values, which, as these results confirm, is particularly challenging for schema matching solutions.
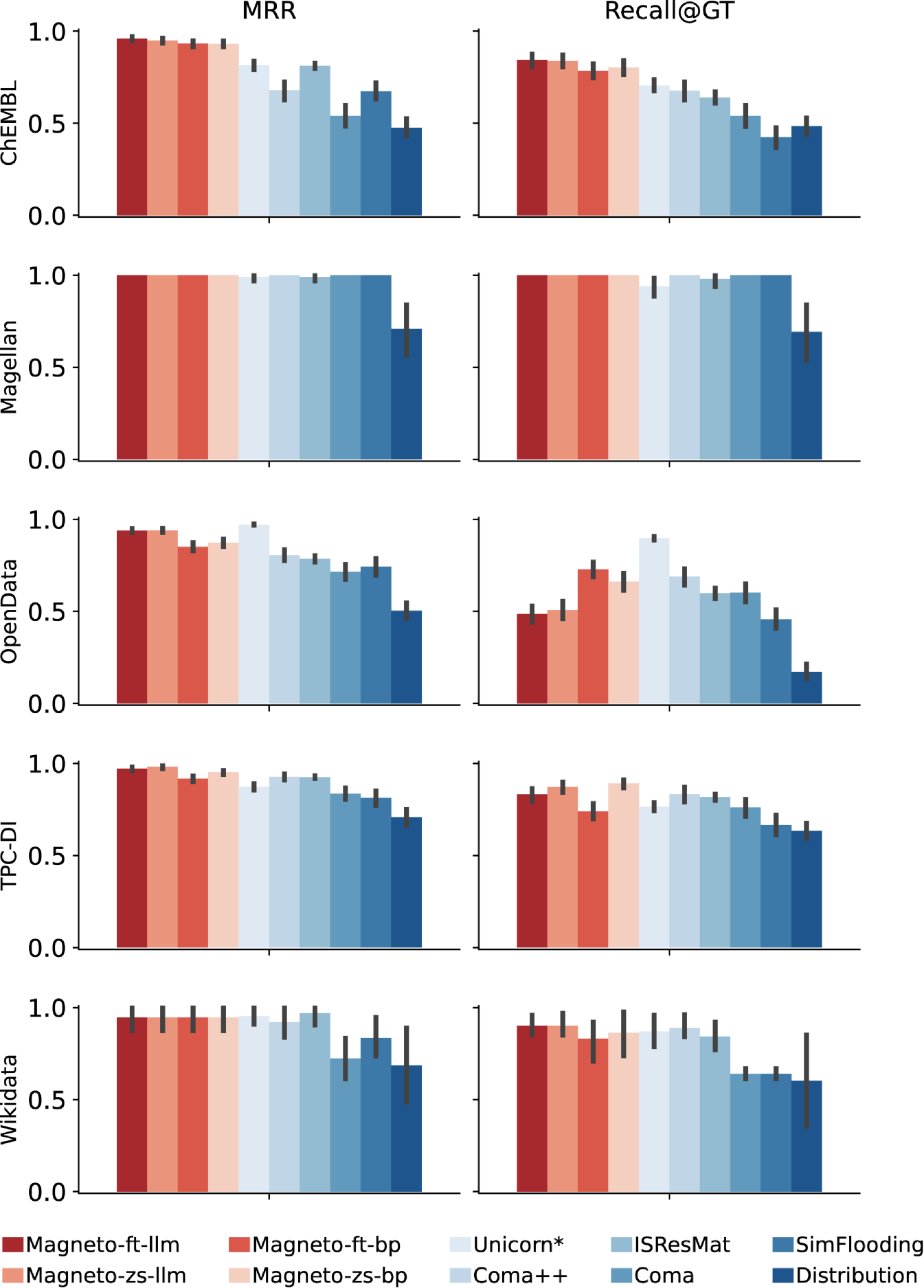
For all Valentine datasets, Magneto performs on par with or better than the baselines. The only exception is Unicorn. However, as previously noted, this can likely be attributed to data leakage: Unicorn is trained using Valentine datasets. This advantage is particularly apparent in datasets like OpenData: Unicorn achieves significantly higher recall than competing methods. Conversely, for the Magellan dataset, which was not used to train Unicorn, Unicorn has lower accuracy than the other methods, including methods that use only basic metadata, which achieve perfect scores.
The high accuracy of the zero-shot configurations of Magneto leaves little room for improvement. Specifically, fine-tuning faces challenges within the Valentine datasets, which predominantly contain lexical rather than semantic matches. For instance, the OpenData dataset includes matches like Gender to Ge and Employer to Em, which are uncommon in practical applications. Despite these obstacles, our LLM-based fine-tuning and re-ranking generally enhance performance. Additionally, we demonstrate in Section 6 that combining simple synthetic data generation techniques with LLM-derived data leads to better performance in this context.
6.3. Scalability Assessment
We compare the runtime of Magneto to those of the top three baselines in accuracy: Unicorn, Coma++, and ISResMat. Since Coma++ does not support GPU execution, we ran this experiment in CPU mode to ensure a fair comparison. We report only the results of Magneto-zs-bp and Magneto-zs-llm since their runtime is similar to Magneto-ft-bp and Magneto-ft-llm, respectively.
For this experiment, we focus on datasets featuring tables with a large number of columns or rows, and we select one source-target pair each from the GDC and OpenData. For the GDC dataset, the source table comprises 179 rows and 153 columns, while the target table contains 4.5k rows and 733 columns. For OpenData, both the source and the target table contain 23k rows and 43 columns. We maintain the input table static and incrementally increase the number of target columns using a random selection. Each execution is repeated 10 times per column number to accommodate randomness. We used a time limit of 2 hours per execution and canceled the executions of any method that exceeded this limit. The results are shown in Figure 8.
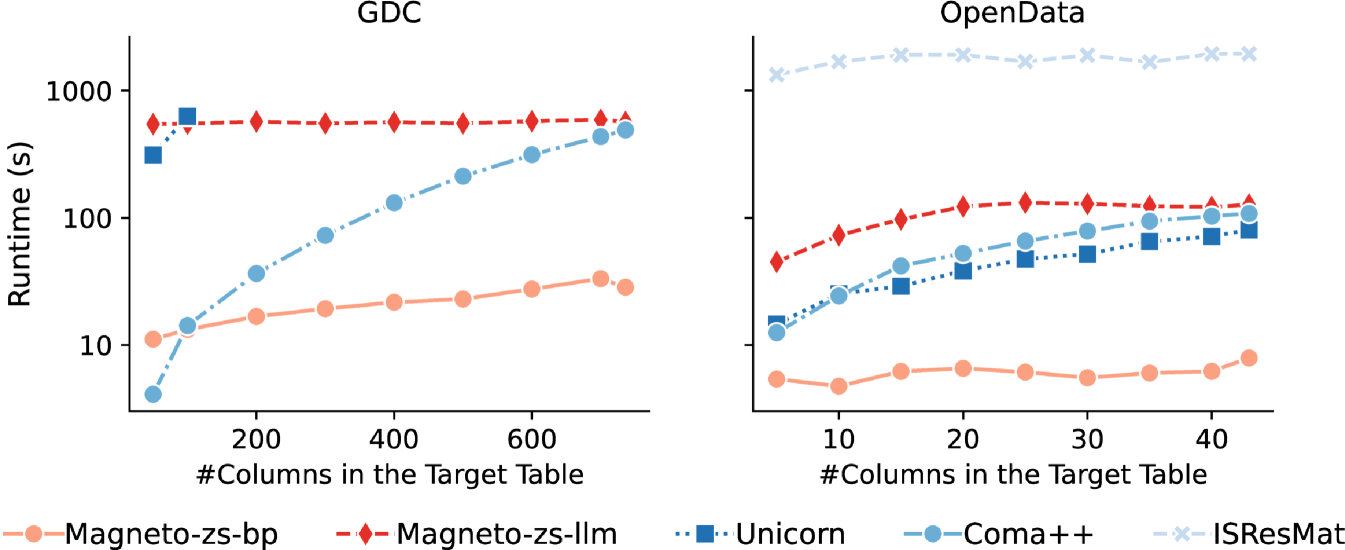
Magneto-zs-bp and Magneto-zs-llm remain stable with increasing table size: the runtime for Magneto-zs-bp grows slightly and Magneto-zs-llm maintains a stable runtime despite its complexity. For GDC, Magneto-zs-bp shows runtimes ranging from 11–33 seconds. Such a low increase in runtime reflects its efficient design, bounded primarily by the embedding computations. As expected, Magneto-zs-llm incurs higher runtimes due to the LLM requests (545–589 seconds), but it shows stability as the number of columns increases since the amount of data sent does not change. The runtimes of Magneto variations are even lower for OpenData: the dataset has fewer columns but many more rows, which Magneto compensates through sampling.
Coma++ shows low runtimes for a small number of columns, but its performance decreases as the number of columns grows. IsResMat and Unicorn exhibit significant scalability challenges, as their runtime grows substantially with the number of columns. IsResMat was not able to complete the execution for GDC, not even for the lowest number of columns, and its runtime for OpenData was orders of magnitude higher than the other methods. Unicorn could only process the initial 100 columns for GDC.
6.4. Ablation of Magneto Components
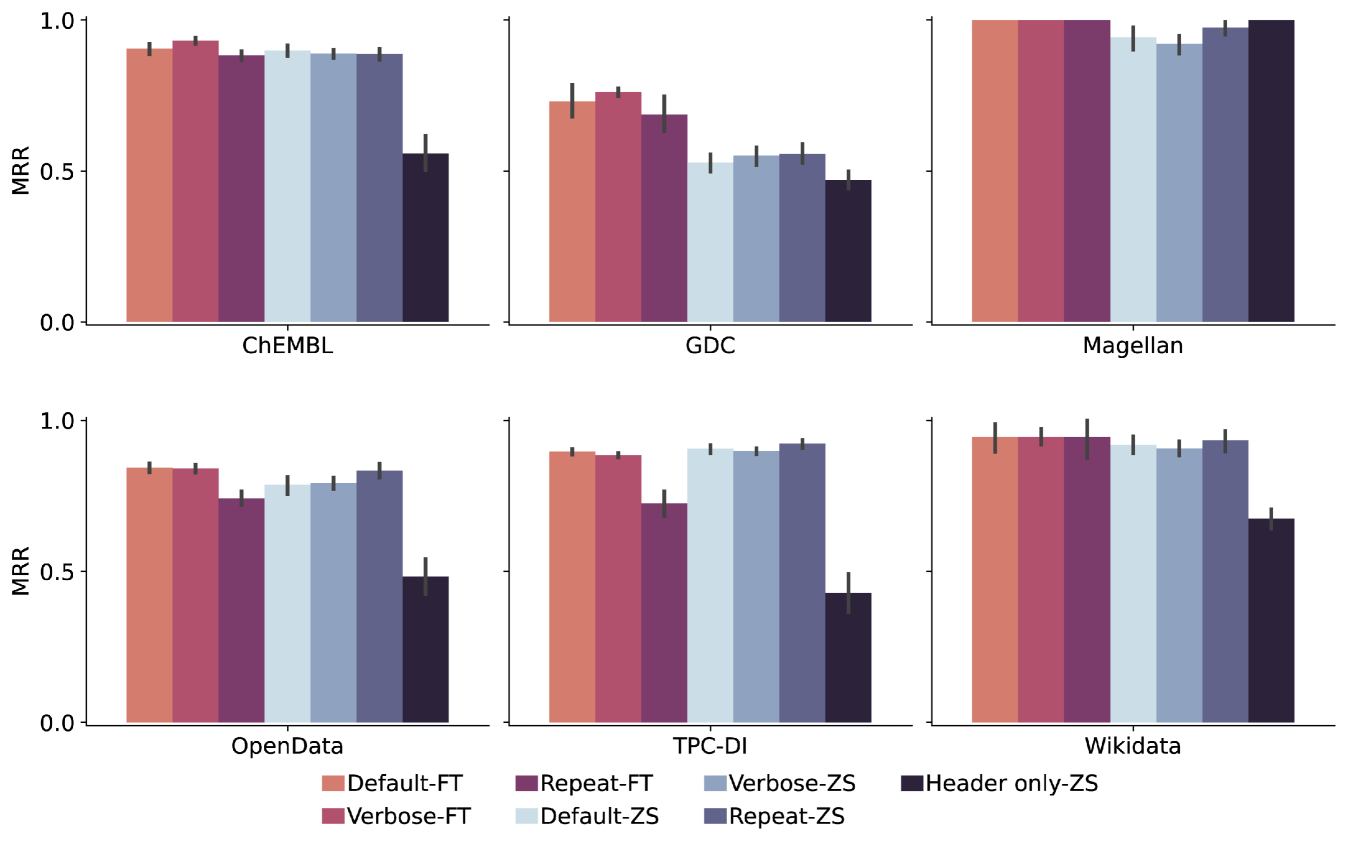
Column Serialization. Figure 9 shows the impact of the serialization methods (Section 3.1) on schema matching performance across all datasets based on MRR. We used the serialization strategies on fine-tuned (FT) and zero-shot (ZS) settings: Default, using , which combines column names with a small sample of values; Verbose, which uses and enriches by incorporating additional instructions; Repeat, which emphasizes header names with . We also include Header only-ZS, which uses only column headers without values, serving as a baseline.
In zero-shot settings, consistently outperforms other methods, demonstrating the effectiveness of emphasizing the column header and raising its cumulative attention in the zero-shot regime. While under-performs in zero-shot settings, it excels after fine-tuning because the prefixes act as learnable anchors that guide the model to separate and re-weight heterogeneous components (name, type, values) during representation learning, highlighting the benefits of integrating semantic details when domain-specific training is applied. The Header only-ZS approach shows the lowest performance in all datasets, confirming that column headers alone are insufficient in complex scenarios.
The results further support our assumption that fine-tuning can greatly improve the performance gains on the GDC dataset. This improvement is likely related to the requirement for external knowledge, which we capture with the LLM-derived training data.
Value Sampling. We compare the effectiveness our proposed Priority Sampling (Section 3.1) against the following sampling methods: Coordinated, a variation of Priority that excludes frequency weights; Weighted and Frequency, variations of Priority without coordination but with value frequencies – Frequency uses the most frequent values and Weighted uses weighted sampling based on these frequencies; Random uses basic random sampling.
We used GDC and the three synthetic datasets from Valentine; we exclude Wikidata and Magellan since as all methods already perform well on them. As the results in Table 4 show, Magneto-zs-bp with , the best zero-shot setting configuration, Priority, generally outperforms other methods in both Recall@GT and MRR, and sometimes is a close second. Priority Sampling prioritizes frequently occurring values and enhances the likelihood of sampling similar values across columns, a beneficial feature for schema matching. The top three techniques—including Priority and its ablations—perform comparably well, also representing viable choices.
Synthetic Data Generation. We evaluate our llm-augfor SLM fine-tuning against two other methods: struct-aug, which incorporates row shuffling, sampling, and column name perturbation, and mixed-aug, which combines these methods. We used the optimal settings from serialization studies, , on Magneto-ft-bp and Magneto-ft-llm. We assess MRR for both methods and Recall@GT for Magneto-ft-bp, as Recall@GT for Magneto-ft-llm is largely influenced by the reranker instead of the fine-tuned candidate retriever. This analysis focuses on the four datasets that have the potential for improvement. Table 5 shows that llm-augand mixed-aug outperform simple perturbations on the GDC dataset. However, on Valentine datasets, LLM-generated data has limited impact due to sparse, uncommon matches. Still, the strong GDC results highlight the effectiveness of our fine-tuning for real data.
| Sampling Method | GDC | ChEMBL | OpenData | TPC-DI |
| Recall@GT | ||||
| Priority | 0.344±0.081 | 0.620±0.264 | 0.543±0.294 | 0.726±0.174 |
| Coordinated | 0.336±0.069 | 0.601±0.260 | 0.506±0.292 | 0.675±0.224 |
| Weighted | 0.342±0.070 | 0.603±0.266 | 0.497±0.291 | 0.643±0.210 |
| Frequency | 0.332±0.062 | 0.525±0.296 | 0.526±0.300 | 0.692±0.196 |
| Random | 0.334±0.075 | 0.572±0.268 | 0.489±0.282 | 0.665±0.197 |
| MRR | ||||
| Priority | 0.591±0.094 | 0.900±0.103 | 0.847±0.200 | 0.948±0.082 |
| Coordinated | 0.586±0.106 | 0.902±0.105 | 0.837±0.196 | 0.937±0.087 |
| Weighted | 0.599±0.099 | 0.888±0.104 | 0.823±0.212 | 0.930±0.091 |
| Frequency | 0.577±0.104 | 0.851±0.163 | 0.833±0.202 | 0.886±0.130 |
| Random | 0.579±0.096 | 0.885±0.106 | 0.830±0.195 | 0.918±0.101 |
| Data Generation | GDC | ChEMBL | OpenData | TPC-DI |
| Recall@GT for Magneto-ft-bp | ||||
| llm-aug | 0.414±0.106 | 0.785±0.263 | 0.729±0.271 | 0.740±0.298 |
| mixed-aug | 0.438±0.085 | 0.774±0.273 | 0.743±0.255 | 0.763±0.255 |
| struct-aug | 0.418±0.099 | 0.764±0.282 | 0.711±0.271 | 0.773±0.261 |
| MRR for Magneto-ft-bp | ||||
| llm-aug | 0.754±0.093 | 0.932±0.103 | 0.851±0.152 | 0.917±0.100 |
| mixed-aug | 0.761±0.071 | 0.931±0.101 | 0.841±0.162 | 0.885±0.125 |
| struct-aug | 0.731±0.099 | 0.927±0.103 | 0.817±0.181 | 0.905±0.117 |
| MRR for Magneto-ft-llm | ||||
| llm-aug | 0.866±0.083 | 0.960±0.089 | 0.939±0.080 | 0.971±0.079 |
| mixed-aug | 0.830±0.077 | 0.949±0.109 | 0.927±0.096 | 0.965±0.096 |
| struct-aug | 0.798±0.116 | 0.955±0.097 | 0.917±0.105 | 0.969±0.081 |
Model Ablations. We extended our ablation studies to include RoBERTa (Liu, 2019) and E5 (Wang et al., 2022) (SLMs), and LLaMA3.3-70B (Dubey et al., 2024) (LLM). Table 6 shows the results. Regarding SLMs the default MPNet outperforms other SLMs in most variations of Magneto. In general, the serialization and score-based prompt proposed are robust. While dominates most zero-shot settings, and yield even higher gains after fine-tuning, which is consistent with our findings in Figure 9. The tested models benefit from the same prompting framework, which confirms that Magneto performs consistently using different models with a robust cost-accuracy tradeoff. For LLMs, LLaMA3.3-70B surpasses GPT-4o-mini (MRR=0.837 vs. 0.815), confirming that larger LLMs enhance reranking. Combining LLaMA3.3-70B with fine-tuned MPNet achieves the best overall performance (MRR=0.860), reinforcing the value of having a pipeline that performs retrieval and reranking.
| Model | zs-bp | ft-bp | zs-gpt4o-mini | ft-gpt4o-mini | zs-llama3.3-70b | ft-llama3.3-70b |
| MPNet () | 0.6560.082 / 0.3570.093 | 0.7400.089 / 0.4000.094 | 0.7750.102 / 0.4120.115 | 0.8460.104 / 0.5370.162 | 0.7720.101 / 0.3390.155 | 0.8600.088 / 0.4230.170 |
| MPNet () | 0.6270.119 / 0.3410.114 | 0.7610.071 / 0.4380.085 | 0.7580.118 / 0.4690.131 | 0.8300.766 / 0.4790.154 | 0.7760.098 / 0.3230.153 | 0.8380.121 / 0.4460.185 |
| MPNet () | 0.7310.086 / 0.3750.108 | 0.7010.110 / 0.1720.103 | 0.8080.095 / 0.4300.131 | 0.8200.117 / 0.5240.104 | 0.8280.096 / 0.3580.117 | 0.8190.102 / 0.4190.138 |
| RoBERTa () | 0.6310.105 / 0.3360.081 | 0.7030.120 / 0.3780.107 | 0.7340.119 / 0.3810.088 | 0.7840.131 / 0.4900.149 | 0.7550.098 / 0.2920.119 | 0.8150.121 / 0.4320.164 |
| RoBERTa () | 0.6210.063 / 0.3500.083 | 0.6920.114 / 0.3670.112 | 0.7430.097 / 0.4000.099 | 0.8210.092 / 0.5110.160 | 0.7510.075 / 0.3240.140 | 0.8540.101 / 0.4240.147 |
| RoBERTa () | 0.7100.103 / 0.3730.097 | 0.6810.096 / 0.3680.111 | 0.7940.097 / 0.4340.173 | 0.7740.101 / 0.5040.161 | 0.8140.088 / 0.3980.161 | 0.8100.090 / 0.4280.165 |
| E5 () | 0.6230.104 / 0.3290.105 | 0.7290.081 / 0.4060.094 | 0.7380.144 / 0.3560.144 | 0.8320.080 / 0.4800.117 | 0.7560.120 / 0.4050.090 | 0.8570.076 / 0.3880.112 |
| E5 () | 0.6020.094 / 0.3220.102 | 0.6920.114 / 0.3670.112 | 0.7450.131 / 0.3820.135 | 0.8210.092 / 0.5110.160 | 0.7460.100 / 0.3270.100 | 0.8380.121 / 0.4460.185 |
| E5 () | 0.7150.120 / 0.3720.100 | 0.7010.110 / 0.3720.103 | 0.7970.121 / 0.3790.112 | 0.8130.113 / 0.5020.154 | 0.8120.118 / 0.3250.144 | 0.8100.090 / 0.4280.165 |
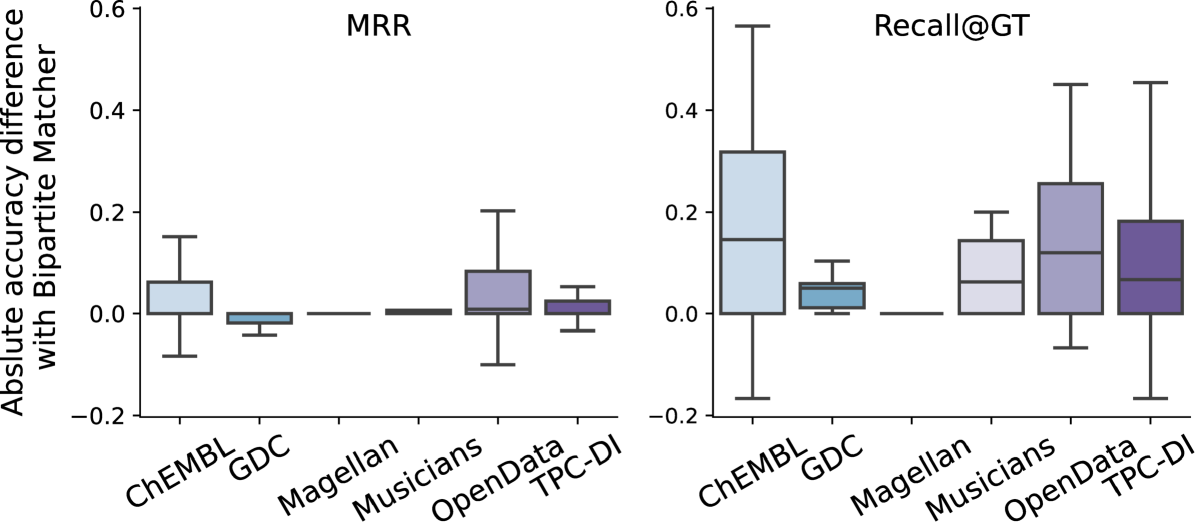
Match Reranker. We first examine the impact of using the bipartite strategy (Section 2.2). Figure 10 shows the increase/decrease in accuracy from Magneto-zs, which directly returns the retriever matches, to Magneto-zs-bp. Note that the bipartite approach can significantly improve Recall@GT. All datasets benefit from the technique at some point, except Magellan, whose scores are already very high. The absolute increase can reach nearly 0.6. The improvements for MRR are lower but still sizable for ChEMBL and OpenData.
We also evaluate: (1) improvements of the LLM-based reranker over the bipartite method, (2) performance variations with more candidates sent to the LLM, and (3) outcomes when bypassing the SLM retrieval stage. We focus on the GDC dataset, noted for its high column count, using zero-shot settings for the retriever. Figure 11 presents accuracy and runtime for different numbers of candidates () processed by GPT-4o-mini in Magneto-zs-llm, comparing it against the use of LLaMA3.3-70B (Llama) and Magneto-zs-bp (BP).
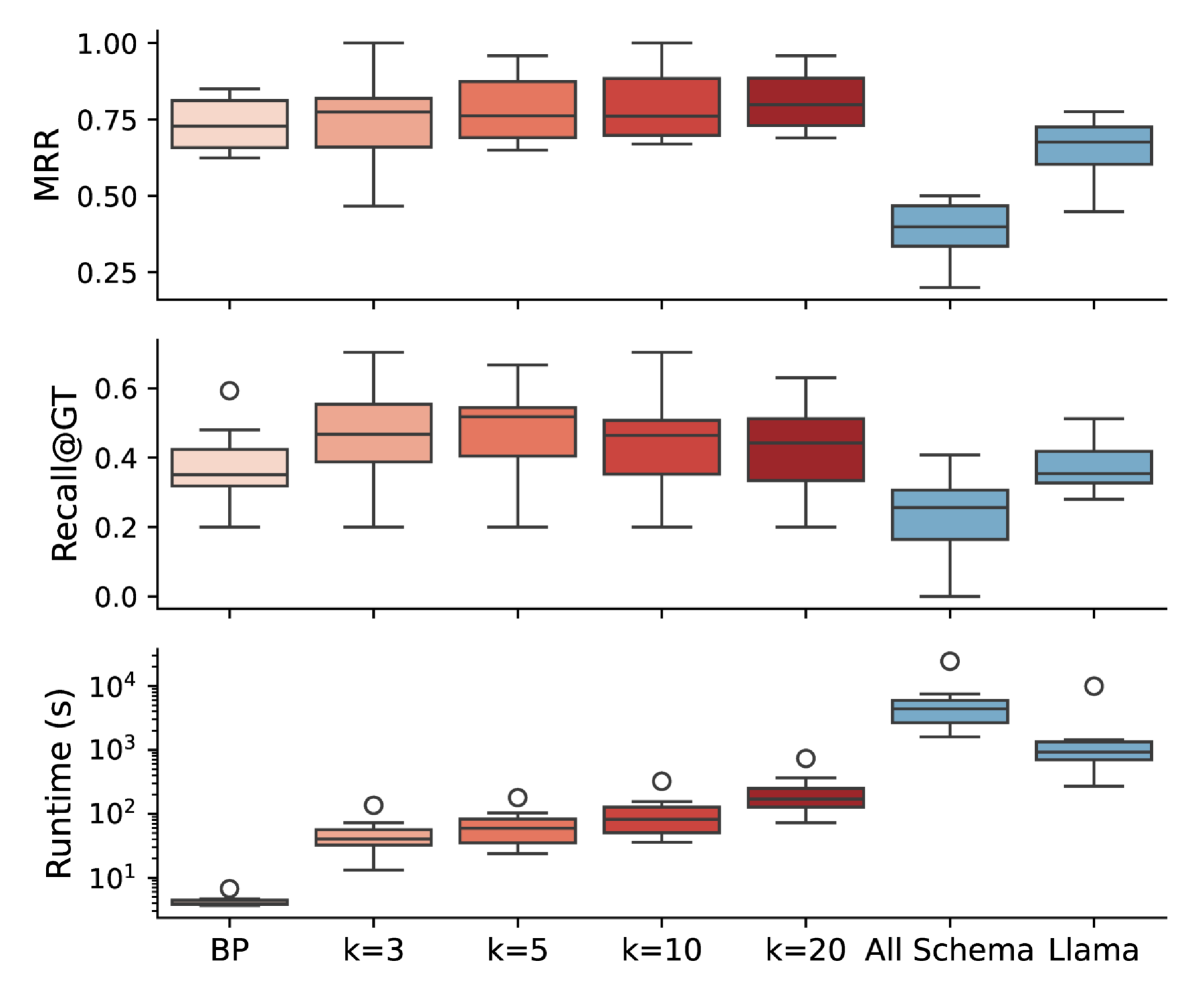
The LLM-based re-ranking approach improves over the bipartite. For example, at , we observe a 6.7% improvement in MRR over the bipartite baseline, rising from 0.731 to 0.780. Recall@GT shows even more promising gains, from 0.375 to 0.475—a 26.7% increase. Increasing enhances MRR, although non-forwarded candidates score lower, reducing Recall@GT which considers all pairs.
When all schemas sent to the LLM reranker, both GPT-4o-mini (All Schema) and LLaMA3.3-70B (Llama), the performance is lower than all Magneto variations. Furthermore, runtime increases with column count and is substantial when reranking all schemas, reaching over 6,000 seconds per table for GPT-4o due to LLM API latency. In contrast, when the SLM-based retriever is used, the runtime remains practical. We used for our other experiments, as it balances good MRR, Recall@GT, and runtime across GDC and other datasets. These results demonstrate the effectiveness of combining SLMs and LLMs for schema matching.
7. Related Work
This section discusses schema and ontology matching approaches related to our work.
Traditional Methods. A straightforward approach for schema matching is detecting overlap in column names (Madhavan et al., 2001; Melnik et al., 2002; Do and Rahm, 2002) and overlap in column values (Aumueller et al., 2005; Zhang et al., 2011). Some incorporate relaxations when measuring overlaps, such as accounting for syntactic and semantic similarities between column names and values (Madhavan et al., 2001; Aumueller et al., 2005). Others also consider factors like data type relevance and value distribution (Aumueller et al., 2005; Zhang et al., 2011). Among these, the COMA algorithm stands out for integrating the most strategies and weighting their outputs to achieve better accuracy (Do and Rahm, 2002; Aumueller et al., 2005), often remaining competitive even against more recent approaches (Koutras et al., 2021). However, these approaches often struggle to capture complex relationships and deeper semantics within datasets (Khatiwada et al., 2023).
Small Language Model-Driven Methods. Methods based on small language models (SLMs) usually use embeddings to encode and compare column data (Cappuzzo et al., 2020). Contrastive learning can improve an SLM’s ability to distinguish matching and non-matching column pairs (Fan et al., 2023; Cong et al., 2023; Zhang et al., 2023), while synthetic tabular data generation can help models to improve without ground truth (Fan et al., 2023; Du et al., 2024). Among these methods, ISResMat customizes pre-trained models for dataset-specific adaptation, generating training pairs from table fragments, and applying pairwise comparison losses to refine matching accuracy (Du et al., 2024). Unicorn, a general matching model, employs contrastive learning and a Mixture-of-Experts (MoE) layer within its architecture to discern matches but relies on supervised training (Tu et al., 2023).
Large Language Model-Driven Methods. Recent works have leveraged large language models (LLMs) for various aspects of tabular data management, predominantly focusing on single-table tasks (Kayali et al., 2024; Hegselmann et al., 2023). Some studies discuss LLM applications for schema matching and highlight the potential of LLMs for this task (Li et al., 2024a; Feuer et al., 2024). However, these approaches often rely solely on prompting strategies—either fine-tuned or zero-shot—which suffer from scalability issues and high computational costs (Li et al., 2024a; Parciak et al., 2024; Fang et al., 2024; Feng et al., 2024). Sheetrit et al. (2024) and Zhang et al. (2024) utilize zero-shot pre-trained LLMs for schema matching. Sheetrit et al. (2024) addresses multiple-table matching, whereas our work focuses on two-table schema matching, particularly for tables with numerous columns. Zhang et al. (2024) uses rule-based feature extraction and trains an XGBoost classifier with gold data, a supervised approach distinct from our unsupervised method. Xu et al. (2024) proposed manually-derived rules to guide LLMs during the matching process and incorporated external knowledge to deal with hallucinations. Parciak et al. (2024) explored various prompting strategies for matching source attributes to a target schema. Note that the evaluation of these methods only take into account table/attribute names and descriptions (Xu et al., 2024; Parciak et al., 2024)—unlike Magneto, they do not consider values.
Ontology Matching. Ontology matching (OM) is related to schema matching (SM) but focuses on identifying semantic correspondences between elements (e.g., classes and properties) across different ontologies expressed in languages like OWL and RDF/XML (Euzenat and Shvaiko, 2013). Some OM approaches use heuristics, rule-based methods (Euzenat and Shvaiko, 2013; Hertling et al., 2019), structural similarity, linguistics, and domain-specific resources (Jiménez-Ruiz et al., 2012; Harrow et al., 2017). To facilitate the matching of domain-specific terminology, semi-automatic annotation using vocabularies and ontologies has been used to enrich schema labels (Sorrentino et al., 2011; Beneventano et al., 2015). Recent OM approaches leverage LLMs and, like Magneto, use a two-phase matching process (Babaei Giglou et al., 2025; Hertling and Paulheim, 2023). OLaLa uses embeddings for candidate matches and LLM evaluation with natural language conversion (Hertling and Paulheim, 2023), while LLMs4OM employs RAG to extract and classify concept similarities (Babaei Giglou et al., 2025). Unlike these methods that prompt per candidate pair, Magneto efficiently generates a ranked list for each input column.
Adapting OM methods to SM is not straightforward: OM approaches target structured relationships rather than flat tabular data, may disregard the similarity of column values, and require format conversions that can impact performance. We compared Magneto against two OM systems, LogMap (Jiménez-Ruiz et al., 2012) and LLMs4OM (Babaei Giglou et al., 2025), by treating tables as classes and columns as properties. They underperformed SM-based baselines by a large margin. Nonetheless, given their shared goals and challenges, combining OM and SM techniques is an interesting direction for future work.
8. Conclusions and Future work
We proposed Magneto, a framework that leverages small and large language models to derive schema matching strategies that generalize across domains and balance accuracy and runtime tradeoffs. We introduced a new benchmark that captures some of the complexities in biomedical data integration and presents new challenges for schema matching. With a detailed experimental evaluation, including comparisons against state-of-the-art methods and ablations, we demonstrate the effectiveness of Magneto and our design choices.
There are several directions for our future work. Magneto ’s accuracy depends on SLM retrieval—if the correct match is missing from the top-, reranking cannot recover it. We plan to explore hybrid retrieval strategies and training SLMs on large, multi-domain datasets. To address limitations in underrepresented domains, we also aim to integrate external knowledge for better zero-shot performance. Reranking quality can vary due to prompt dependence—a known limitation of instruction-tuned LLMs (Feuer et al., 2024). Future work may leverage prompt tuning frameworks like DSPy (Khattab et al., 2024) for systematic, task-aware optimization. While LLM-based reranking improves accuracy, it introduces cost overheads. Future work could optimize reranking with smaller, efficient LLMs.
Acknowledgments
This work was supported by NSF awards IIS-2106888 and OAC-2411221, the DARPA ASKEM program Agreement No. HR0011262087, and the ARPA-H BDF program. The views, opinions, and findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the DARPA, ARPA-H, the U.S. Government, or NSF. We thank our collaborators from the NYU School of Medicine, David Fenyo, Wenke Liu, and Sarah Keegan, for sharing their experience in biomedical data integration and contributing to the GDC benchmark. We also thank Roque Lopez for his contributions to the code.
References
- (1)
- Alserafi et al. (2020) Ayman Alserafi, Alberto Abelló, Oscar Romero, and Toon Calders. 2020. Keeping the Data Lake in Form: Proximity Mining for Pre-Filtering Schema Matching. ACM Transactions on Information Systems (TOIS) 38, 3 (2020), 1–30. https://doi.org/10.1145/3388870
- ARPA-H (2024) ARPA-H. 2024. Biomedical Data Fabric (BDF) Toolbox. https://arpa-h.gov/research-and-funding/programs/arpa-h-bdf-toolbox. Accessed: 2024-11-13.
- Aumueller et al. (2005) David Aumueller, Hong-Hai Do, Sabine Massmann, and Erhard Rahm. 2005. Schema and ontology matching with COMA++. In In Proceedings of the ACM International Conference on Management of Data (SIGMOD). 906–908.
- Babaei Giglou et al. (2025) Hamed Babaei Giglou, Jennifer D’Souza, Felix Engel, and Sören Auer. 2025. LLMs4OM: Matching Ontologies with Large Language Models. In The Semantic Web: ESWC 2024 Satellite Events, Albert Meroño Peñuela, Oscar Corcho, Paul Groth, Elena Simperl, Valentina Tamma, Andrea Giovanni Nuzzolese, Maria Poveda-Villalón, Marta Sabou, Valentina Presutti, Irene Celino, Artem Revenko, Joe Raad, Bruno Sartini, and Pasquale Lisena (Eds.). 25–35.
- Badaro et al. (2023) Gilbert Badaro, Mohammed Saeed, and Paolo Papotti. 2023. Transformers for Tabular Data Representation: A Survey of Models and Applications. Transactions of the Association for Computational Linguistics 11 (2023), 227–249. https://doi.org/10.1162/tacl_a_00544
- Beneventano et al. (2015) Domenico Beneventano, Sonia Bergamaschi, Serena Sorrentino, Maurizio Vincini, and Fabio Benedetti. 2015. Semantic annotation of the CEREALAB database by the AGROVOC linked dataset. Ecological Informatics 26 (2015), 119–126. https://doi.org/10.1016/j.ecoinf.2014.07.002
- Brown et al. (2020) Tom Brown, Benjamin Mann, Nick Ryder, Melanie Subbiah, Jared D Kaplan, Prafulla Dhariwal, Arvind Neelakantan, Pranav Shyam, Girish Sastry, Amanda Askell, et al. 2020. Language models are few-shot learners. Advances in neural information processing systems 33 (2020), 1877–1901.
- Cafarella et al. (2009) Michael J. Cafarella, Alon Halevy, and Nodira Khoussainova. 2009. Data integration for the relational web. Proceedings of the VLDB Endowment (PVLDB) 2, 1 (2009), 1090–1101.
- Cao et al. (2021) Liwei Cao, Chen Huang, Daniel Cui Zhou, Yingwei Hu, T Mamie Lih, Sara R Savage, Karsten Krug, David J Clark, Michael Schnaubelt, Lijun Chen, et al. 2021. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 184, 19 (2021), 5031–5052.
- Cappuzzo et al. (2020) Riccardo Cappuzzo, Paolo Papotti, and Saravanan Thirumuruganathan. 2020. Creating embeddings of heterogeneous relational datasets for data integration tasks. In In Proceedings of the ACM International Conference on Management of Data (SIGMOD). 1335–1349.
- Castro Fernandez et al. (2018) Raul Castro Fernandez, Essam Mansour, Abdulhakim A. Qahtan, Ahmed Elmagarmid, Ihab Ilyas, Samuel Madden, Mourad Ouzzani, Michael Stonebraker, and Nan Tang. 2018. Seeping Semantics: Linking Datasets Using Word Embeddings for Data Discovery. In 2018 IEEE 34th International Conference on Data Engineering (ICDE). 989–1000.
- Cheng et al. (2024) Cindy Cheng, Luca Messerschmidt, Isaac Bravo, Marco Waldbauer, Rohan Bhavikatti, Caress Schenk, Vanja Grujic, Tim Model, Robert Kubinec, and Joan Barceló. 2024. A general primer for data harmonization. Scientific data 11, 1 (2024), 152.
- Clark et al. (2019) David J Clark, Saravana M Dhanasekaran, Francesca Petralia, Jianbo Pan, Xiaoyu Song, Yingwei Hu, Felipe da Veiga Leprevost, Boris Reva, Tung-Shing M Lih, Hui-Yin Chang, et al. 2019. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179, 4 (2019), 964–983.
- Cong et al. (2023) Tianji Cong, Fatemeh Nargesian, and HV Jagadish. 2023. Pylon: Semantic Table Union Search in Data Lakes. arXiv preprint arXiv:2301.04901.
- cptac (2024) cptac 2024. Clinical Proteomic Tumor Analysis Consortium (CPTAC). https://proteomics.cancer.gov/programs/cptac. Accessed: 2024-11-13.
- Crouse (2016) David F. Crouse. 2016. On implementing 2D rectangular assignment algorithms. IEEE Trans. Aerospace Electron. Systems 52, 4 (2016), 1679–1696. https://doi.org/10.1109/TAES.2016.140952
- Daliri et al. (2024) Majid Daliri, Juliana Freire, Christopher Musco, Aécio Santos, and Haoxiang Zhang. 2024. Sampling Methods for Inner Product Sketching. Proceedings of the VLDB Endowment (PVLDB) 17, 9 (2024), 2185–2197.
- Das et al. ([n.d.]) Sanjib Das, AnHai Doan, Paul Suganthan G. C., Chaitanya Gokhale, Pradap Konda, Yash Govind, and Derek Paulsen. [n.d.]. The Magellan Data Repository. https://sites.google.com/site/anhaidgroup/projects/data.
- datagov (2024) datagov 2024. U.S. Government’s Open Data. https://data.gov. Accessed: 2024-11-13.
- Devlin (2018) Jacob Devlin. 2018. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
- Do and Rahm (2002) Hong-Hai Do and Erhard Rahm. 2002. COMA: a system for flexible combination of schema matching approaches. In Proceedings of the International Conference on Very Large Data Bases (VLDB). 610–621.
- Doan et al. (2012) AnHai Doan, Alon Halevy, and Zachary Ives. 2012. Principles of Data Integration (1st ed.). Morgan Kaufmann Publishers Inc., San Francisco, CA, USA.
- Dong and Wang (2024) Haoyu Dong and Zhiruo Wang. 2024. Large Language Models for Tabular Data: Progresses and Future Directions. In Proceedings of the International ACM Conference on Research and Development in Information Retrieval (SIGIR). 2997–3000. https://doi.org/10.1145/3626772.3661384
- Dong et al. (2022) Qingxiu Dong, Lei Li, Damai Dai, Ce Zheng, Jingyuan Ma, Rui Li, Heming Xia, Jingjing Xu, Zhiyong Wu, Tianyu Liu, et al. 2022. A survey on in-context learning. arXiv preprint arXiv:2301.00234.
- Dong et al. (2023) Yuyang Dong, Chuan Xiao, Takuma Nozawa, Masafumi Enomoto, and Masafumi Oyamada. 2023. DeepJoin: Joinable Table Discovery with Pre-Trained Language Models. Proceedings of the VLDB Endowment (PVLDB) 16, 10 (2023), 2458–2470.
- Dou et al. (2020) Yongchao Dou, Emily A Kawaler, Daniel Cui Zhou, Marina A Gritsenko, Chen Huang, Lili Blumenberg, Alla Karpova, Vladislav A Petyuk, Sara R Savage, Shankha Satpathy, et al. 2020. Proteogenomic characterization of endometrial carcinoma. Cell 180, 4 (2020), 729–748.
- Du et al. (2024) Xingyu Du, Gongsheng Yuan, Sai Wu, Gang Chen, and Peng Lu. 2024. In Situ Neural Relational Schema Matcher. In IEEE International Conference on Data Engineering (ICDE). IEEE, 138–150.
- Dubey et al. (2024) Abhimanyu Dubey, Abhinav Jauhri, Abhinav Pandey, Abhishek Kadian, Ahmad Al-Dahle, Aiesha Letman, Akhil Mathur, Alan Schelten, Amy Yang, Angela Fan, et al. 2024. The llama 3 herd of models. arXiv preprint arXiv:2407.21783.
- European Organization For Nuclear Research and OpenAIRE ([n.d.]) European Organization For Nuclear Research and OpenAIRE. [n.d.]. Zenodo. https://www.zenodo.org. Accessed: 2024-11-13.
- Euzenat and Shvaiko (2013) Jrme Euzenat and Pavel Shvaiko. 2013. Ontology Matching (2nd ed.). Springer Publishing Company, Incorporated.
- Fan et al. (2023) Grace Fan, Jin Wang, Yuliang Li, Dan Zhang, and Renée J. Miller. 2023. Semantics-Aware Dataset Discovery from Data Lakes with Contextualized Column-Based Representation Learning. Proceedings of the VLDB Endowment (PVLDB) 16, 7 (2023), 1726–1739.
- Fang et al. (2024) Xi Fang, Weijie Xu, Fiona Anting Tan, Ziqing Hu, Jiani Zhang, Yanjun Qi, Srinivasan H. Sengamedu, and Christos Faloutsos. 2024. Large Language Models (LLMs) on Tabular Data: Prediction, Generation, and Understanding - A Survey. Transactions on Machine Learning Research 2024 (2024). https://openreview.net/forum?id=IZnrCGF9WI
- Feng et al. (2024) Longyu Feng, Huahang Li, and Chen Jason Zhang. 2024. Cost-Aware Uncertainty Reduction in Schema Matching with GPT-4: The Prompt-Matcher Framework. arXiv preprint arXiv:2408.14507 (2024).
- Feuer et al. (2024) Benjamin Feuer, Yurong Liu, Chinmay Hegde, and Juliana Freire. 2024. ArcheType: A Novel Framework for Open-Source Column Type Annotation Using Large Language Models. Proceedings of the VLDB Endowment (PVLDB) 17, 9 (2024), 2279–2292.
- Gal (2006) Avigdor Gal. 2006. Managing uncertainty in schema matching with top-k schema mappings. Springer-Verlag, Berlin, Heidelberg, 90–114.
- Gillette et al. (2020) Michael A Gillette, Shankha Satpathy, Song Cao, Saravana M Dhanasekaran, Suhas V Vasaikar, Karsten Krug, Francesca Petralia, Yize Li, Wen-Wei Liang, Boris Reva, et al. 2020. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 182, 1 (2020), 200–225.
- Harrow et al. (2017) Ian Harrow, Ernesto Jiménez-Ruiz, Andrea Splendiani, Martin Romacker, Peter Woollard, Scott Markel, Yasmin Alam-Faruque, Martin Koch, James Malone, and Arild Waaler. 2017. Matching disease and phenotype ontologies in the ontology alignment evaluation initiative. J. Biomed. Semant. 8, 1 (2017), 55:1–55:13. https://doi.org/10.1186/S13326-017-0162-9
- Heath et al. (2021) Allison P Heath, Vincent Ferretti, Stuti Agrawal, Maksim An, James C Angelakos, Renuka Arya, Rosita Bajari, Bilal Baqar, Justin HB Barnowski, Jeffrey Burt, et al. 2021. The NCI genomic data commons. Nature genetics 53, 3 (2021), 257–262.
- Hegselmann et al. (2023) Stefan Hegselmann, Alejandro Buendia, Hunter Lang, Monica Agrawal, Xiaoyi Jiang, and David Sontag. 2023. Tabllm: Few-shot classification of tabular data with large language models. In International Conference on Artificial Intelligence and Statistics. PMLR, 5549–5581.
- Hermans et al. (2017) Alexander Hermans, Lucas Beyer, and Bastian Leibe. 2017. In defense of the triplet loss for person re-identification. arXiv preprint arXiv:1703.07737.
- Hertling and Paulheim (2023) Sven Hertling and Heiko Paulheim. 2023. OLaLa: Ontology Matching with Large Language Models. In Proceedings of the Knowledge Capture Conference (K-CAP) (Pensacola, FL, USA). 131–139. https://doi.org/10.1145/3587259.3627571
- Hertling et al. (2019) Sven Hertling, Jan Portisch, and Heiko Paulheim. 2019. MELT - Matching EvaLuation Toolkit. In Semantic Systems. The Power of AI and Knowledge Graphs - International Conference (SEMANTiCS) (Lecture Notes in Computer Science), Vol. 11702. Springer, 231–245. https://doi.org/10.1007/978-3-030-33220-4_17
- Huang et al. (2021) Chen Huang, Lijun Chen, Sara R Savage, Rodrigo Vargas Eguez, Yongchao Dou, Yize Li, Felipe da Veiga Leprevost, Eric J Jaehnig, Jonathan T Lei, Bo Wen, et al. 2021. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer cell 39, 3 (2021), 361–379.
- Jiménez-Ruiz et al. (2012) Ernesto Jiménez-Ruiz, Bernardo Cuenca Grau, Yujiao Zhou, and Ian Horrocks. 2012. Large-scale Interactive Ontology Matching: Algorithms and Implementation. In European Conference on Artificial Intelligence. https://api.semanticscholar.org/CorpusID:1618779
- Kayali et al. (2024) Moe Kayali, Anton Lykov, Ilias Fountalis, Nikolaos Vasiloglou, Dan Olteanu, and Dan Suciu. 2024. Chorus: Foundation Models for Unified Data Discovery and Exploration. Proceedings of the VLDB Endowment (PVLDB) 17, 8 (2024), 2104–2114.
- Khatiwada et al. (2023) Aamod Khatiwada, Grace Fan, Roee Shraga, Zixuan Chen, Wolfgang Gatterbauer, Renée J. Miller, and Mirek Riedewald. 2023. SANTOS: Relationship-based Semantic Table Union Search. Proceedings of the VLDB Endowment (PVLDB) 1, 1, Article 9 (2023), 25 pages.
- Khattab et al. (2024) Omar Khattab, Arnav Singhvi, Paridhi Maheshwari, Zhiyuan Zhang, Keshav Santhanam, Sri Vardhamanan, Saiful Haq, Ashutosh Sharma, Thomas T. Joshi, Hanna Moazam, Heather Miller, Matei Zaharia, and Christopher Potts. 2024. DSPy: Compiling Declarative Language Model Calls into Self-Improving Pipelines. In International Conference on Learning Representations (ICLR).
- Koutras et al. (2021) Christos Koutras, George Siachamis, Andra Ionescu, Kyriakos Psarakis, Jerry Brons, Marios Fragkoulis, Christoph Lofi, Angela Bonifati, and Asterios Katsifodimos. 2021. Valentine: Evaluating matching techniques for dataset discovery. In IEEE International Conference on Data Engineering (ICDE). IEEE, 468–479.
- Krug et al. (2020) Karsten Krug, Eric J Jaehnig, Shankha Satpathy, Lili Blumenberg, Alla Karpova, Meenakshi Anurag, George Miles, Philipp Mertins, Yifat Geffen, Lauren C Tang, et al. 2020. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell 183, 5 (2020), 1436–1456.
- Li et al. (2024a) Peng Li, Yeye He, Dror Yashar, Weiwei Cui, Song Ge, Haidong Zhang, Danielle Rifinski Fainman, Dongmei Zhang, and Surajit Chaudhuri. 2024a. Table-GPT: Table Fine-tuned GPT for Diverse Table Tasks. Proc. ACM Manag. Data 2, 3, Article 176 (May 2024), 28 pages. https://doi.org/10.1145/3654979
- Li et al. (2024b) Tianle Li, Ge Zhang, Quy Duc Do, Xiang Yue, and Wenhu Chen. 2024b. Long-context llms struggle with long in-context learning. arXiv preprint arXiv:2404.02060 (2024).
- Li et al. (2023) Yize Li, Yongchao Dou, Felipe Da Veiga Leprevost, Yifat Geffen, Anna P Calinawan, François Aguet, Yo Akiyama, Shankara Anand, Chet Birger, Song Cao, et al. 2023. Proteogenomic data and resources for pan-cancer analysis. Cancer Cell 41, 8 (2023), 1397–1406.
- Liu (2019) Yinhan Liu. 2019. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692 364 (2019).
- Madhavan et al. (2001) Jayant Madhavan, Philip A. Bernstein, and Erhard Rahm. 2001. Generic Schema Matching with Cupid. In Proceedings of the International Conference on Very Large Data Bases (VLDB). 49–58.
- McDermott et al. (2020) Jason E McDermott, Osama A Arshad, Vladislav A Petyuk, Yi Fu, Marina A Gritsenko, Therese R Clauss, Ronald J Moore, Athena A Schepmoes, Rui Zhao, Matthew E Monroe, et al. 2020. Proteogenomic characterization of ovarian HGSC implicates mitotic kinases, replication stress in observed chromosomal instability. Cell reports medicine 1, 1 (2020).
- Melnik et al. (2002) Sergey Melnik, Hector Garcia-Molina, and Erhard Rahm. 2002. Similarity flooding: A versatile graph matching algorithm and its application to schema matching. In IEEE International Conference on Data Engineering (ICDE). IEEE, 117–128.
- Mertins et al. (2016) Philipp Mertins, D. R. Mani, Kelly V. Ruggles, Michael A. Gillette, Karl R. Clauser, Pei Wang, Xianlong Wang, Jana W. Qiao, Song Cao, Francesca Petralia, Emily Kawaler, Filip Mundt, Karsten Krug, Zhidong Tu, Jonathan T. Lei, Michael L. Gatza, Matthew Wilkerson, Charles M. Perou, Venkata Yellapantula, Kuan-lin Huang, Chenwei Lin, Michael D. McLellan, Ping Yan, Sherri R. Davies, R. Reid Townsend, Steven J. Skates, Jing Wang, Bing Zhang, Christopher R. Kinsinger, Mehdi Mesri, Henry Rodriguez, Li Ding, Amanda G. Paulovich, David Fenyö, Matthew J. Ellis, Steven A. Carr, and NCI CPTAC. 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 7605 (2016), 55–62. https://doi.org/10.1038/nature18003
- Miller (2018) Renée J. Miller. 2018. Open data integration. Proceedings of the VLDB Endowment (PVLDB) 11, 12 (2018), 2130–2139.
- Narayan et al. (2022) Avanika Narayan, Ines Chami, Laurel Orr, and Christopher Ré. 2022. Can Foundation Models Wrangle Your Data? Proceedings of the VLDB Endowment (PVLDB) 16, 4 (2022), 738–746.
- National Cancer Institute (2024a) National Cancer Institute. 2024a. Genomics Data Commons (GDC). https://gdc.cancer.gov. Accessed: 2024-11-13.
- National Cancer Institute (2024b) National Cancer Institute. 2024b. Search - GDC Docs. https://docs.gdc.cancer.gov/Data_Dictionary/gdcmvs/. Accessed: 2024-12-01.
- National Institutes of Health (2020) National Institutes of Health. 2020. NIH Data Management and Sharing Policy. Online. https://sharing.nih.gov/data-management-and-sharing-policy Available at: https://sharing.nih.gov/data-management-and-sharing-policy.
- Nature (2024) Nature. 2024. Reporting standards and availability of data, materials, code and protocols. https://www.nature.com/nature-portfolio/editorial-policies/reporting-standards. Accessed: 2024-11-13.
- of New York (2024) City of New York. 2024. NYC Open Data Portal. https://opendata.cityofnewyork.us.
- Parciak et al. (2024) Marcel Parciak, Brecht Vandevoort, Frank Neven, Liesbet M Peeters, and Stijn Vansummeren. 2024. Schema Matching with Large Language Models: an Experimental Study. arXiv preprint arXiv:2407.11852 (2024).
- pdc (2023) pdc 2023. Proteomic Data Commons. https://proteomic.datacommons.cancer.gov/pdc.
- Radwan et al. (2009) Ahmed Radwan, Lucian Popa, Ioana R. Stanoi, and Akmal Younis. 2009. Top-k generation of integrated schemas based on directed and weighted correspondences. In In Proceedings of the ACM International Conference on Management of Data (SIGMOD). 641–654.
- Santos et al. (2025) Aécio Santos, Eden Wu, Roque Lopez, Sarah Keegan, Eduardo Pena, Wenke Liu, Yurong Liu, David Fenyo, and Juliana Freire. 2025. GDC-SM: The GDC Schema Matching Benchmark. https://doi.org/10.5281/zenodo.14963588
- Satpathy et al. (2021) Shankha Satpathy, Karsten Krug, Pierre M Jean Beltran, Sara R Savage, Francesca Petralia, Chandan Kumar-Sinha, Yongchao Dou, Boris Reva, M Harry Kane, Shayan C Avanessian, et al. 2021. A proteogenomic portrait of lung squamous cell carcinoma. Cell 184, 16 (2021), 4348–4371.
- Schroff et al. (2015) Florian Schroff, Dmitry Kalenichenko, and James Philbin. 2015. Facenet: A unified embedding for face recognition and clustering. In Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). 815–823.
- Schütze et al. (2008) Hinrich Schütze, Christopher D Manning, and Prabhakar Raghavan. 2008. Introduction to information retrieval. Vol. 39. Cambridge University Press Cambridge.
- Science (2024) Science 2024. Science Journals: Editorial Policies. https://www.science.org/content/page/science-journals-editorial-policies. Accessed: 2024-11-13.
- Sheetrit et al. (2024) Eitam Sheetrit, Menachem Brief, Moshik Mishaeli, and Oren Elisha. 2024. ReMatch: Retrieval Enhanced Schema Matching with LLMs. arXiv preprint arXiv:2403.01567 (2024).
- Shraga et al. (2020) Roee Shraga, Avigdor Gal, and Haggai Roitman. 2020. ADnEV: cross-domain schema matching using deep similarity matrix adjustment and evaluation. Proceedings of the VLDB Endowment (PVLDB) 13, 9 (2020), 1401–1415.
- Song et al. (2020) Kaitao Song, Xu Tan, Tao Qin, Jianfeng Lu, and Tie-Yan Liu. 2020. Mpnet: Masked and permuted pre-training for language understanding. Advances in neural information processing systems (NeurIPS) 33 (2020), 16857–16867.
- Sorrentino et al. (2011) Serena Sorrentino, Sonia Bergamaschi, and Maciej Gawinecki. 2011. NORMS: An automatic tool to perform schema label normalization. In 2002 IEEE International Conference on Data Engineering (ICDE). 1344–1347.
- Suhara et al. (2022) Yoshihiko Suhara, Jinfeng Li, Yuliang Li, Dan Zhang, Çağatay Demiralp, Chen Chen, and Wang-Chiew Tan. 2022. Annotating columns with pre-trained language models. In Proceedings of the International Conference on Management of Data (SIGMOD). 1493–1503.
- The bdi-kit contributors (2024) The bdi-kit contributors. 2024. bdi-kit - A Python toolkit for data harmonization. https://bdi-kit.readthedocs.io/. Accessed: 2024-12-01.
- Tu et al. (2023) Jianhong Tu, Ju Fan, Nan Tang, Peng Wang, Guoliang Li, Xiaoyong Du, Xiaofeng Jia, and Song Gao. 2023. Unicorn: A unified multi-tasking model for supporting matching tasks in data integration. Proceedings of the ACM on Management of Data 1, 1 (2023), 1–26.
- ukbio (2024) ukbio 2024. UK Biobank. https://www.ukbiobank.ac.uk. Accessed: 2024-11-13.
- Vasaikar et al. (2019) Suhas Vasaikar, Chen Huang, Xiaojing Wang, Vladislav A Petyuk, Sara R Savage, Bo Wen, Yongchao Dou, Yun Zhang, Zhiao Shi, Osama A Arshad, et al. 2019. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 177, 4 (2019), 1035–1049.
- Vaswani et al. (2017) Ashish Vaswani, Noam Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N Gomez, Łukasz Kaiser, and Illia Polosukhin. 2017. Attention is all you need. Advances in neural information processing systems 30 (2017).
- Voorhees et al. (1999) Ellen M Voorhees et al. 1999. The trec-8 question answering track report.. In Trec, Vol. 99. 77–82.
- Wang et al. (2022) Liang Wang, Nan Yang, Xiaolong Huang, Binxing Jiao, Linjun Yang, Daxin Jiang, Rangan Majumder, and Furu Wei. 2022. Text embeddings by weakly-supervised contrastive pre-training. arXiv preprint arXiv:2212.03533 (2022).
- Wang et al. (2021) Liang-Bo Wang, Alla Karpova, Marina A Gritsenko, Jennifer E Kyle, Song Cao, Yize Li, Dmitry Rykunov, Antonio Colaprico, Joseph H Rothstein, Runyu Hong, et al. 2021. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer cell 39, 4 (2021), 509–528.
- White et al. (2023) J. White, Q. Fu, S. Hays, M. Sandborn, C. Olea, H. Gilbert, A. Elnashar, J. Spencer-Smith, and D. C. Schmidt. 2023. A Prompt Pattern Catalog to Enhance Prompt Engineering with ChatGPT. (2023). arXiv:arXiv:2302.11382 [cs.CL]
- Xu et al. (2024) Yongqin Xu, Huan Li, Ke Chen, and Lidan Shou. 2024. KcMF: A Knowledge-compliant Framework for Schema and Entity Matching with Fine-tuning-free LLMs. arXiv preprint arXiv:2410.12480 (2024).
- Yin et al. (2020) Pengcheng Yin, Graham Neubig, Wen-tau Yih, and Sebastian Riedel. 2020. TaBERT: Pretraining for joint understanding of textual and tabular data. arXiv preprint arXiv:2005.08314 (2020).
- Zhang et al. (2011) Meihui Zhang, Marios Hadjieleftheriou, Beng Chin Ooi, Cecilia M. Procopiuc, and Divesh Srivastava. 2011. Automatic discovery of attributes in relational databases. In In Proceedings of the ACM International Conference on Management of Data (SIGMOD). 109–120.
- Zhang et al. (2024) Yu Zhang, Mei Di, Haozheng Luo, Chenwei Xu, and Richard Tzong-Han Tsai. 2024. SMUTF: Schema Matching Using Generative Tags and Hybrid Features. arXiv preprint arXiv:2402.01685 (2024).
- Zhang et al. (2023) Yunjia Zhang, Avrilia Floratou, Joyce Cahoon, Subru Krishnan, Andreas C Müller, Dalitso Banda, Fotis Psallidas, and Jignesh M Patel. 2023. Schema matching using pre-trained language models. In IEEE International Conference on Data Engineering (ICDE). IEEE, 1558–1571.