RANCOR: Non-Linear Image Registration with Total Variation Regularization
Abstract
Optimization techniques have been widely used in deformable registration, allowing for the incorporation of similarity metrics with regularization mechanisms. These regularization mechanisms are designed to mitigate the effects of trivial solutions to ill-posed registration problems and to otherwise ensure the resulting deformation fields are well-behaved. This paper introduces a novel deformable registration algorithm, RANCOR, which uses iterative convexification to address deformable registration problems under total-variation regularization. Initial comparative results against four state-of-the-art registration algorithms are presented using the Internet Brain Segmentation Repository (IBSR) database.
keywords:
Deformable Image Registration, Total Variation Regularization, Brain, Convex Relaxation, GPGPU1 Introduction
Registration is the systematic spatial deformation of one medical image as to allign it with another, either in the same or a different modality. Although registration of images common to a single patient can rely largely on rigid transformations, registration between patient images, common in techniques such as atlas construction or atlas-based segmentation, have relied on highly non-linear (NLR) deformations often in the absence of highly detectable and localizable landmarks. Non-linear registration aims to address these problems, using a similiarity metric to judge the quality of the alignment after deformation, and a regularization mechanism to ensure that the deformation field avoid trivial and otherwise undesirable components such as gaps or singularities. Non-linear registration is indeed a challenging problem with many competing facets. Our algorithm is intended to provide an additional option that could facilitate atlas building and segmentation techniques.
Many components of our deformable registration algorithm display a large amount of inherent parallelism between image voxels. Such algorithms have been of growing interest to the medical imaging community because of the ability to implement them on commercially avialable general purpose graphics processing units (GPGPUs) to dramatically improve their speed and computational efficiency for both registration[1] and segmentation problems [2, 3, 4].
1.1 Contributions
We propose a novel non-linear RegistrAtioN via COnvex Relaxation (RANCOR) algorithm that allows for the combination of any pointwise error metric (such as the sum of absolute intensity differences (SAD) for intra-modality registration, and mutual information (MI) [5, 6] or modality independent neighbourhood descriptors (MIND) [7] for inter-modality registration) while regularizing the deformation field by its total variation. This algorithm is implemented on GPGPU to ensure high performance.
1.2 Previous studies
Recent surveys provide a good overview of existing NLR registration methods [8, 9] and we would like to emphasize the study performed by Klein et al. [10], where 14 NLR registration algorithms were compared across four open brain image databases. We will compare our proposed method against the four highest ranked methods identified in [10]:
Advanced Normalization Tools (ANTs): The Symmetric Normalization (SyN) NLR method in [11] uses a multi-resolution scheme to enforce a bi-directional diffeomorphism while maximizing a cross-correlation metric. It has been shown in several open challenges [12, 10, 13] to outperform well established methods. SyN regularizes the deformation field through Gaussian smoothing and enforcing transformation symmetry.
Image Registration Roolkit (IRTK): The well-known Fast Free-Form deformations (F3D) method in [14] defines a lattice of equally spaced control points over the target image and, by moving each point, locally modifies the deformation field. Normalized mutual information combined with a cubic b-spline bending energy is used as the objective function. Its multi-resolution implementation employs coarsely-to-finely spaced lattices and Gaussian smoothing.
Automatic Registration Toolbox (ART): [15] presents a homeomorphic NLR method using normalized cross-correlation as similarity metric in a multi-resolution framework. The deformation field is regularized via median and low-pass Gaussian filtering.
Statistical Parametric Mapping DARTEL Toolbox (SPM_D): The DARTEL algorithm presented in [16] employs a static finite difference model of a velocity field. The flow field is considered as a member of the Lie algebra, which is exponentiated to produce a deformation inherently enforcing a diffeomorphism. It is implemented in a recursive, multi-resolution manner.
2 Methods
In this section, we propose a multi-scale dual optimization based method to estimate the non-linear deformation field , bewteen two given images and , which explores the minimization of the variational optical-flow energy function:
| (1) |
where the function term stands for a dissimilarity measure of the two input images and under deformation by , and gives the regularization function to single out a smooth deformation field. In this paper, we use the sum of absolute intensity differences (SAD):
| (2) |
as a simple similarity metric for two input images from the same modality. The proposed framework can also be directly adapted for more advanced image dissimilarity measures designed for registration between different modalities.
A regularization term, , is often incorporated to make the minimization problem (1) well-posed. Otherwise, minimizing the image dissimilarity function can result in trivial or infinite solutions. We consider the total variation of the deformation field as the regularization term:
| (3) |
The expected non-convexity of and , makes it challenging to directly minimize (1), even with convex regularization. To address this issue, we introduce an incremental convexification approach, which lends itself to a standard coarse-to-fine framework and allows for a more global perspective and avoiding local optima by capturing large deformations.
In Section 2.1, we develop the multi-scale optimization framework, developing a sequence of related minimization problems. Each of these problems are solved through a new non-smooth Gauss-Newton (GN) approach introduced in Section 2.2. which employs a novel sequential convexification and dual optimization procedure.
2.1 Coarse-to-Fine Optimization Framework
The first stage in our approach is the construction of the image pyramid. Let … be the -level pyramid representation of from the coarsest resolution to the finest resolution , and … the -level coarse-to-fine pyramid representation of . The optimization process is started from the coarsest level, , which extracts the deformation field between and such that:
| (4) |
The vector field gives the optimal deformation field at the coarsest scale. It is warped to the next finer-resolved level, , to compute the optimal finer-level defomration field . The process is repeated, obtaining the deformation field … at each level sequentially.
Second, at each resolution level , , we compute an incremental deformation field based on the two image functions and , where is warped by the deformation field computed at the previous resolution level , i.e.
| (5) |
Clearly, the optimization problem (4) can be viewed as the special case of (5), i.e. for , we define and . Therefore, the proposed coarse-to-fine optimization framework sequentially explores the minimization of (5) at each image resolution level, from the coarsest to the finest .
2.2 Sequential Convexification and Dual Optimization
Now we consider the optimization problem (5) for each image resolution level . Given the highly non-linear function in (5), we introduce a sequential linearization and convexification procedure for this challenging non-linear optimization problem (5). This results in a series of incremental warping steps in which each step approximates an update of the deformation field , until the updated deformation is sufficiently small, i.e., it iterates through the following sequence of convex minimization steps until convergence is attained:
-
•
Initialize and let ;
-
•
At the iteration, define the deformation field as
and compute the update deformation to by minimizing the following convex energy function:
(6) where
and .
-
•
Let and repeat the second step till the new update is small enough. Then, we have the total incremental deformation field at the image resolution level as:
These steps can be viewed as a non-smooth GN method for the non-linear optimization problem (5), in contrast to the classical GN method proposed in [17]. Moreover, the -norm and the convex regularization term , (6) results in a convex optimization problem. The non-smooth -norm from (6) provides more robustness in practice than the conventional smooth -norm used in the classical GN method.
Solving the convex minimization problem (6) is the most essential step in the proposed algorithmic framework The introduced primal-dual variational analysis not only provides an equivalent dual formulation to the optimization problem (6) but also derives an efficient solution algorithm. First, we simplify the expression of the convex problem (6) as:
| (7) |
where represents the deformation field. Through variational analysis, we can derive an equivalent dual model to (7):
Proposition 2.1
The proof is given in Appendix A.
As shown in Appendix A, each component of the deformation field works as the optimal multiplier functions to their respective constraints, (9). Therfore, the energy function of the primal-dual model (24) is exactly the Lagrangian function to the dual model (8):
where and the linear functions , , are defined in (8) and (9) respectively. We can now derive an efficient duality-based Lagrangian augmented algorithm based on the modern convex optimization theories (see [18, 19, 20] for details), using the augmented Lagrangian function:
| (10) |
where is a positive constant and the additional quadratic penalty function is applied to ensure the functions (9) vanish. Our proposed duality-based optimization algorithm is:
-
•
Set the initial values of , and , and let .
-
•
Fix and , optimize by
(11) generating the convex minimization problem:
(12) where () is computed from the fixed variables and . is computed by threshholding:
(13) where
-
•
Fixing and , optimize by
(14) which amounts to three convex minimization problems:
(15) where is computed from the fixed variables and . Hence, , , can be approximated by a gradient-projection step corresponding to (22).
-
•
Once and are obtained, update by
(16) -
•
Increment and iterate until converged, i.e.
(17) where is a chosen small positive parameter ().
3 Experiments
3.1 Image Database
The image data constisted of an open multi-center T1w MRI dataset with corresponding manual segmentations, the Internet Brain Segmentation Repository (IBSR) database, totalling 18 labeled image volumes at 1.5T available on www.mindboggle.info in a pre-processed form with labeling protocols and transforms into MNI space.
The experiments were performed in a pair-wise manner. For each image in the database, seventeen registrations were performed using the chosen image as the reference image and one of the remaining as the floating image. Thus, our experiment consisted of 306 registration problems in total.
3.2 Initialization & Pre-processing
Prior to registration, all images were skull stripped by constructing brain masks from manual labels using morphological operations [10] and then affinely registered using the FMRIB Software Library’s (FSL) FLIRT package [21] into the space of the MNI152_T1_1mm_brain. These affine transformations were made available on www.mindboggle.info and used to initialize the NLR algorithms. This guarantees that the same initialization is used for the algorithms in [10] and allows for quantitative comparisons. As a pre-processing step, both affinely registered images were robustly normalized to zero mean and standard deviation units to ensure a constant regularization weight could be used.
3.3 Implementation & Parameter Tuning
The proposed NLR method was implemented in MATLAB (Natick, MA) using the Compute Unified Device Architecture (CUDA) (NVIDIA, Santa Clara, CA) for GPGPU computing. Each level in the coarse-to-fine framework constists of multiple warps invoking the proposed GPGPU accelerated regularization algorithm. Parameter tuning of the regularization weight was done on two randomly picked dataset pairs similar to the tuning in [10]. All other parameters, such as the number of levels (), the number of warps () and the maximum number of iterations () were determined heuristically on a single image volume not used in this study. Table 1 contains all set parameter values.
| Method | ||||
|---|---|---|---|---|
| RANCOR TVR | 0.30 | 3 | 4 | 220 |
| All parameters were kept constant across all experiments. | ||||
3.4 Evaluation Metric
To compare our registration method against other NLR registration algorithms, we used the target overlap (TO) as a regional metric:
| (18) |
where is the floating image, the reference image, and a labeled region, as indicated in [10]. This parallels our motivation of using NLR registration to port segmentation labels to incoming datasets, and takes advantage of the manual segmentations providing in the IBSR database.
Results were considered significant if the probability of making a type I error was less than 1% (). For this purpose, we employed a series of two-tailed, pairwise Student’s t-test.
4 Results
4.1 Run times
The experiments were conducted on a Ubuntu 12.04 (64-bit) desktop machine with 144 GB memory and an NVIDIA Tesla C2060 (6 GB memory) graphics card. The maximum run times for the MATLAB code including pre-processing, optimization, and GPGPU enhanced regularization are given in Table 2.
| TVR | TVR | TVR | Total run time | |
| RANCOR TVR | 0.25 | 1.76 | 13.58 | 76.76 |
4.2 Accuracy
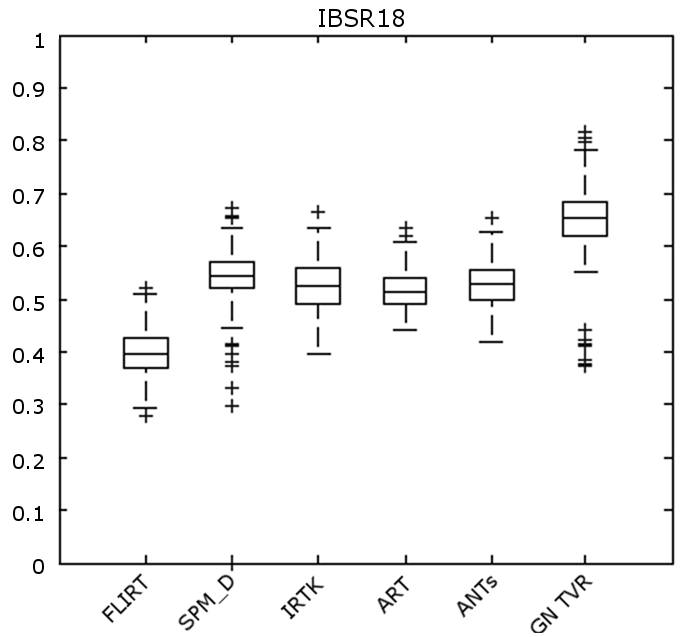
Figure 1 shows boxplots of the TO accuracy for each of the registration methods. The results were averaged across all regions. Numerical results are provided in Table 3.
| IBSR18 | |
|---|---|
| FLIRT | |
| SPM_D | |
| IRTK | |
| ART | |
| Syn | |
| RANCOR TVR |
5 Discussion
We proposed a novel GPGPU-accelerated deformation field regularization method, total variation-based regularization, for NLR registration. This method was implemented within a coarse-to-fine optimization framework and compared on an open and publicly available database, IBSR. We employed the same initialization, tuning conditions, and evaluation scripts to quantitatively compare the proposed methods against four well-known NLR methods. Further, we numerically state the accuracy metrics allowing for direct comparison.
The proposed method significantly outperformed the comparative methods in terms of TO (). We want to note that both the proposed methods employed the simplest and most non-robust similarity metric, SAD, while SPM_D, IRTK, ART and SyN use advanced metrics (see [10]). The choice of similarity metric was intentionally chosen for these experiments to demontrate the potential of the proposed method without better similarity metrics or an advanced optimizer (i.e. a Levenberg-–Marquardt optimizer as used in SPM_D [16]).
5.1 Future directions
The current RANCOR framework can be seen as a basic method to be extended over time, under the same open science credo, that allowed us to readily and quantitatively compare well-known open methods using public databases. As the current framework cannot currently guarantee diffeomorphic deformations, the next step is to enforce such constraints on the resulting deformation fields. Furthermore, to enable inter-modality NLR, we will implement and test commonly used advanced similarity metrics, such as normalized mutual-information, normalized cross-correlation, or more recently developed methods, such as the of the MIND descriptor [7]. Since command-line tools, such as the compared open NLR methods are needed for large-scale data analysis, RANCOR will be definitely included into such a package and, as a matter of course, be made available to the community.
Additionally, we plan to extend our evaluation to include the RANCOR algorithm under an -norm regularization as a substitute for total variation as used in Sun et al[22]. Such evaluation would allow for rigorous comparisons to be made by isolating the regularization mechanism.
6 Conclusions
We proposed a novel GPGPU-accelerated registration algorithm that optimizes any pointwise similarity metric and total variation regularization within a Gauss-Newton optimization framework. This algorithm was then evaluated against the four highest ranking non-linear registration algorithms according to [10] on an open image database. We intend to provide our implementation back to the community in an open manner.
Acknowledgments
Appendix A Dual Optimization Analysis
Gven the conjugate representation of the absolute function:
| (19) |
we can rewrite the first -norm term of (7) as follows:
| (20) |
Additionally, given in terms of (3), we have
| (21) |
where each dual variable , , has characteristic function of the constraint , :
| (22) |
References
- [1] M. Modat, G. R. Ridgway, Z. A. Taylor, M. Lehmann, J. Barnes, D. J. Hawkes, N. C. Fox, and S. Ourselin, “Fast free-form deformation using graphics processing units,” Computer methods and programs in biomedicine, vol. 98, no. 3, pp. 278–284, 2010.
- [2] M. Rajchl, J. Yuan, E. Ukwatta, and T. Peters, “Fast interactive multi-region cardiac segmentation with linearly ordered labels,” in Biomedical Imaging (ISBI), 2012 9th IEEE International Symposium on. IEEE Conference Publications, 2012, pp. Page–s.
- [3] W. Qiu, J. Yuan, E. Ukwatta, Y. Sun, M. Rajchl, and A. Fenster, “Fast globally optimal segmentation of 3d prostate mri with axial symmetry prior,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013. Springer Berlin Heidelberg, 2013, pp. 198–205.
- [4] M. Rajchl, J. Yuan, J. White, E. Ukwatta, J. Stirrat, C. Nambakhsh, F. Li, and T. Peters, “Interactive hierarchical max-flow segmentation of scar tissue from late-enhancement cardiac mr images,” IEEE Transactions on Medical Imaging, vol. 33, no. 1, pp. 159–172, 2014.
- [5] W. M. Wells III, P. Viola, H. Atsumi, S. Nakajima, and R. Kikinis, “Multi-modal volume registration by maximization of mutual information,” Medical image analysis, vol. 1, no. 1, pp. 35–51, 1996.
- [6] P. Rogelj, S. Kovačič, and J. C. Gee, “Point similarity measures for non-rigid registration of multi-modal data,” Computer vision and image understanding, vol. 92, no. 1, pp. 112–140, 2003.
- [7] M. P. Heinrich, M. Jenkinson, M. Bhushan, T. Matin, F. V. Gleeson, S. M. Brady, and J. A. Schnabel, “Mind: Modality independent neighbourhood descriptor for multi-modal deformable registration,” Medical Image Analysis, vol. 16, no. 7, pp. 1423–1435, 2012.
- [8] W. Crum, T. Hartkens, and D. Hill, “Non-rigid image registration: theory and practice,” 2004.
- [9] A. Gholipour, N. Kehtarnavaz, R. Briggs, M. Devous, and K. Gopinath, “Brain functional localization: a survey of image registration techniques,” Medical Imaging, IEEE Transactions on, vol. 26, no. 4, pp. 427–451, 2007.
- [10] A. Klein, J. Andersson, B. A. Ardekani, J. Ashburner, B. Avants, M.-C. Chiang, G. E. Christensen, D. L. Collins, J. Gee, P. Hellier et al., “Evaluation of 14 nonlinear deformation algorithms applied to human brain mri registration,” Neuroimage, vol. 46, no. 3, pp. 786–802, 2009.
- [11] B. B. Avants, C. L. Epstein, M. Grossman, and J. C. Gee, “Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain,” Medical image analysis, vol. 12, no. 1, pp. 26–41, 2008.
- [12] K. Farahani, B. Menze, M. Reyes, E. Gerstner, J. Kirby, and J. Kalpathy-Cramer. (2013, Sep.) Multimodal brain tumor segmentation (brats 2013). [Online]. Available: http://martinos.org/qtim/miccai2013/index.html
- [13] K. Murphy, B. Van Ginneken, J. M. Reinhardt, S. Kabus, K. Ding, X. Deng, K. Cao, K. Du, G. E. Christensen, V. Garcia et al., “Evaluation of registration methods on thoracic ct: the empire10 challenge,” Medical Imaging, IEEE Transactions on, vol. 30, no. 11, pp. 1901–1920, 2011.
- [14] D. Rueckert, L. I. Sonoda, C. Hayes, D. L. Hill, M. O. Leach, and D. J. Hawkes, “Nonrigid registration using free-form deformations: application to breast mr images,” Medical Imaging, IEEE Transactions on, vol. 18, no. 8, pp. 712–721, 1999.
- [15] B. A. Ardekani, S. Guckemus, A. Bachman, M. J. Hoptman, M. Wojtaszek, and J. Nierenberg, “Quantitative comparison of algorithms for inter-subject registration of 3d volumetric brain mri scans,” Journal of neuroscience methods, vol. 142, no. 1, pp. 67–76, 2005.
- [16] J. Ashburner, “A fast diffeomorphic image registration algorithm,” Neuroimage, vol. 38, no. 1, pp. 95–113, 2007.
- [17] M. Baust, D. Zikic, N. Navab, and G. Munich, “Diffusion-based regularisation strategies for variational level set segmentation.” in BMVC, 2010, pp. 1–11.
- [18] D. P. Bertsekas, “Nonlinear programming,” 1999.
- [19] J. Yuan, E. Bae, and X.-C. Tai, “A study on continuous max-flow and min-cut approaches,” in Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference on. IEEE, 2010, pp. 2217–2224.
- [20] J. Yuan, E. Bae, X.-C. Tai, and Y. Boykov, “A continuous max-flow approach to potts model,” in Computer Vision–ECCV 2010. Springer, 2010, pp. 379–392.
- [21] M. Jenkinson, P. Bannister, M. Brady, and S. Smith, “Improved optimization for the robust and accurate linear registration and motion correction of brain images,” Neuroimage, vol. 17, no. 2, pp. 825–841, 2002.
- [22] Y. Sun, J. Yuan, M. Rajchl, W. Qiu, C. Romagnoli, and A. Fenster, “Efficient convex optimization approach to 3d non-rigid mr-trus registration,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013. Springer, 2013, pp. 195–202.