Robustness and Games Against Nature in Molecular Programming
Abstract
Matter, especially DNA, is now programmed to carry out useful processes at the nanoscale. As these programs and processes become more complex and their envisioned safety-critical applications approach deployment, it is essential to develop methods for engineering trustworthiness into molecular programs. Some of this can be achieved by adapting existing software engineering methods, but molecular programming also presents new challenges that will require new methods. This paper presents a method for dealing with one such challenge, namely, the difficulty of ascertaining how robust a molecular program is to perturbations of the relative “clock speeds” of its various reactions. The method proposed here is game-theoretic. The robustness of a molecular program is quantified in terms of its ability to win (achieve its original objective) in games against other molecular programs that manipulate its relative clock speeds. This game-theoretic approach is general enough to quantify the security of a molecular program against malicious manipulations of its relative clock speeds. However, this preliminary report focuses on games against nature, games in which the molecular program’s opponent perturbs clock speeds randomly (indifferently) according to the probabilities inherent in chemical kinetics.
1 Introduction
Molecular programming is, at its simplest, computation with DNA. A programmed molecular system is a nanosystem that will execute the algorithmic behavior encoded into it. Examples of programmed DNA systems include neural net simulation, probabilistic switching circuits, nano-robotic walkers, and oscillators [21, 27, 7, 24]. That is, we are programming matter itself when we create a programmed molecular system.
Many of the intended uses of molecular programming are safety-critical, such as biosensors to detect pollutants, diagnostic devices to identify diseases, and nano-robotic walkers to perform targeted drug delivery [9]. Software engineering techniques including goal-oriented requirements modeling [26], risk analyses [14], and probabilistic model checking [15] have recently been extended to the emerging field of molecular programming, in order to aid the development of safe programmed molecular systems [16, 15, 10].
Assuring the robustness of a molecular program needs to occur before such a system is deployed. However, what robustness means for such a system is not well-defined. This, in turn, hinders efforts to determine how robust a particular system is.
The problem addressed by this paper is the difficulty of answering the question, “how robust is this molecular program?” The contribution of the paper is to propose a game-theoretic method by which we can quantitatively evaluate how robust a molecular program is to an opponent’s perturbing the relative clock speeds of its constituent processes (reactions). This sort of robustness is especially important, because the “rate constants” that govern the rates of chemical reactions are notoriously approximate and non-constant in actual laboratory experiments. We formulate this as a game against nature [17, 19], in which nature manipulates clock speeds at random, disinterested in the outcome. Although this approach is general enough to evaluate robustness in the face of a game against a malicious opponent, we focus here on the random perturbations inflicted by an indifferent nature. We illustrate our method on an important consensus algorithm, approximate majority [5]. We thus develop a games-against-nature formalism of robustness and then evaluate it on a small molecular program that computes the approximate majority.
2 Molecular Programs
Molecular programs are typically specified as chemical reaction networks (CRNs) [1, 6], which are roughly equivalent to stochastic Petri nets [8]. These CRNs can then be automatically compiled into DNA strand displacement systems that can be implemented in laboratory experiments [23, 4, 2].
Syntax
We now review the definition of CRNs. We fix a countably infinite set whose elements are called species. We informally regard each species as an abstract type of molecule.
A reaction over a finite set is an ordered triple
where and is the set of functions from into . Given such a reaction , we call the reactant vector of , the product vector of , and the rate constant of . (Since is finite, it is natural to regard elements of as vectors.) The species in the support are the reactants of , and the species in are the products of .
We usually write reactions in a more chemistry-like notation. For example, if , then we write
for the reaction , where , , , , , and . A species satisfying , as in this example, is called a catalyst of the reaction . The net effect of a reaction is the vector . The arity of a reaction is
i.e., its total number of reactants. A chemical reaction network (CRN) is an ordered pair , where is finite and is a finite set of reactions over .
Semantics
In this paper we assign each CRN the operational meaning given by the stochastic mass action semantics (also called the stochastic mass action kinetics) introduced by Gillespie [13]. In this semantics a state of is a vector . For each , the component of is the count of species in the state . A reaction is applicable to state if , i.e., all the reactants of are present in . If is applicable to , then the result of applying to is the state .
The (stochastic mass action) rate of a reaction in a state and volume of solution, which we denote by , was defined and justified by Gillespie [13]. Here we give a single example. Let be a reaction
(The right-hand side RHS does not affect the rate of a reaction.) For brevity, write and . If is applicable to (i.e., if and ), then
Under stochastic mass-action semantics, a CRN functions as a continuous-time Markov chain [22] with state space and, for each , transition rate
The CRN is initialized to some state or distribution over states. When it enters a state , it stays there for a random, real-valued sojourn time before instantaneously executing some reaction and jumping to the state . A trajectory of is thus a sequence of ordered pairs , where is a state of and is the associated sojourn time. The trajectory is finite if it reaches a state to which no reaction is applicable. Otherwise, the trajectory is infinite.
3 CRN Games
To quantify the robustness of a CRN, we consider how its performance might be affected by other CRNs that are present in the same solution. Clearly, this evaluation will depend both on how performance is defined and on what kinds of other CRNs are present. We begin by describing a general game-theoretic framework that allows for any scalar quantification of performance (by defining appropriate utility functions) and arbitrary constraints placed on the other CRNs (by restricting the other players’ strategy spaces). We then discuss the special case where interactions between CRNs are mediated only by catalysts.
An -player CRN game with players is a pair with the following components.
-
1.
is the strategy profile space. To play the game, each player selects a strategy: a CRN from its strategy space , which is a set of CRNs. For convenience we require that . The players’ selected strategies collectively define a strategy profile and a CRN , where and . A state of the game comprises counts of all species in . As in a CRN, a trajectory in this game is a (finite or infinite) sequence of states paired with sojourn times. The space of all trajectories for the game is .
-
2.
is a profile of utility functions , where is the utility player gets from trajectory . In our example below, this utility function is simply a binary indicator for “success,” meaning that the player gets utility 1 from any trajectory that performs a given task correctly and gets utility 0 from all other trajectories. A player representing “nature” is totally indifferent to the game’s outcome and hence gets utility 0 from all trajectories.
Let where is the random vector for ’s initial state, and let be the embedding of in . Then the initial state of is the random vector . Given this initial distribution, the theory of continuous-time Markov chains specifies a probability measure on the set of all trajectories of the CRN [22], which also immediately yields a probability measure on . This allows us to define the function
which is the expected value of player ’s utility when the strategy profile is played.
Robustness
We measure the robustness of a CRN to a profile of other players’ CRNs by comparing player 1’s expected utility playing against those CRNs to its expected utility playing against trivial CRNs. Formally, for any , a CRN is -robust to in game under if
where and is the trivial distribution that assigns probability 1 to the empty vector. This means that the participation of other players using these strategies can decrease player 1’s expected utility by at most a factor of .
Catalytic Games
The very general CRN games that we have defined allow essentially unrestricted interactions among the players’ CRNs. For many purposes, including those of this paper, it is more appropriate to restrict these interactions to those mediated by catalysts.
A catalytic game is one in which each player’s set of species can be written as such that
-
1.
and are disjoint for all , and
-
2.
for all and , we have .
In such a game, each player can affect other players only by altering the counts of their catalysts, and hence only by altering the rates of their reactions.
4 Example
Approximate Majority
In this preliminary report, we use a game against nature to investigate the robustness of a simple chemical reaction network that computes approximate majority.
The task in approximate majority is to design a chemical reaction network with two designated species and and the following objective. Let and be the counts of and , respectively, at time . First, the total population should be constant as varies. Moreover, if and differ by a non-negligible amount, then whichever is larger should eventually “take over.” That is, if , then we should with high probability have (and hence ) for all sufficiently large . Similarly, if , then we should with high probability have for all sufficiently large . If is very close to , then we want one of these “takeovers” to occur with high probability, but it may be either species that takes over.
We investigate the robustness of the approximate majority CRN
of Condon, Hajiaghayi, Kirkpatrick and Manuch [5]. In order to do this in a catalytic game, we replace with the catalyzed CRN
The crucial thing to note here is that, if , , , and are the counts of , , , and at some time, then the rates (“clock speeds”) of the reactions in at this time are and , respectively, while the rates of the reactions in are and , respectively. If and are equal and constant, then is merely a uniformly sped-up version of . However, if and vary randomly, then the relative rates of the reactions in also vary randomly (i.e., the ratio of these rates varies randomly).
In order to test the robustness of to random perturbations of the relative rates of their reactions we thus play the CRN against a random “nature” that varies the initially equal counts and randomly. We model this behavior by the simple CRN
which is “calibrated” by the rate constant .
Simulation
We assessed the robustness of the approximate majority CRN by comparing the performance of the catalyzed CRN in isolation with its performance in the presence of the CRN representing nature randomly perturbing rate constants. We frame this as a game where the utility of the approximate majority player is given by its success frequency.
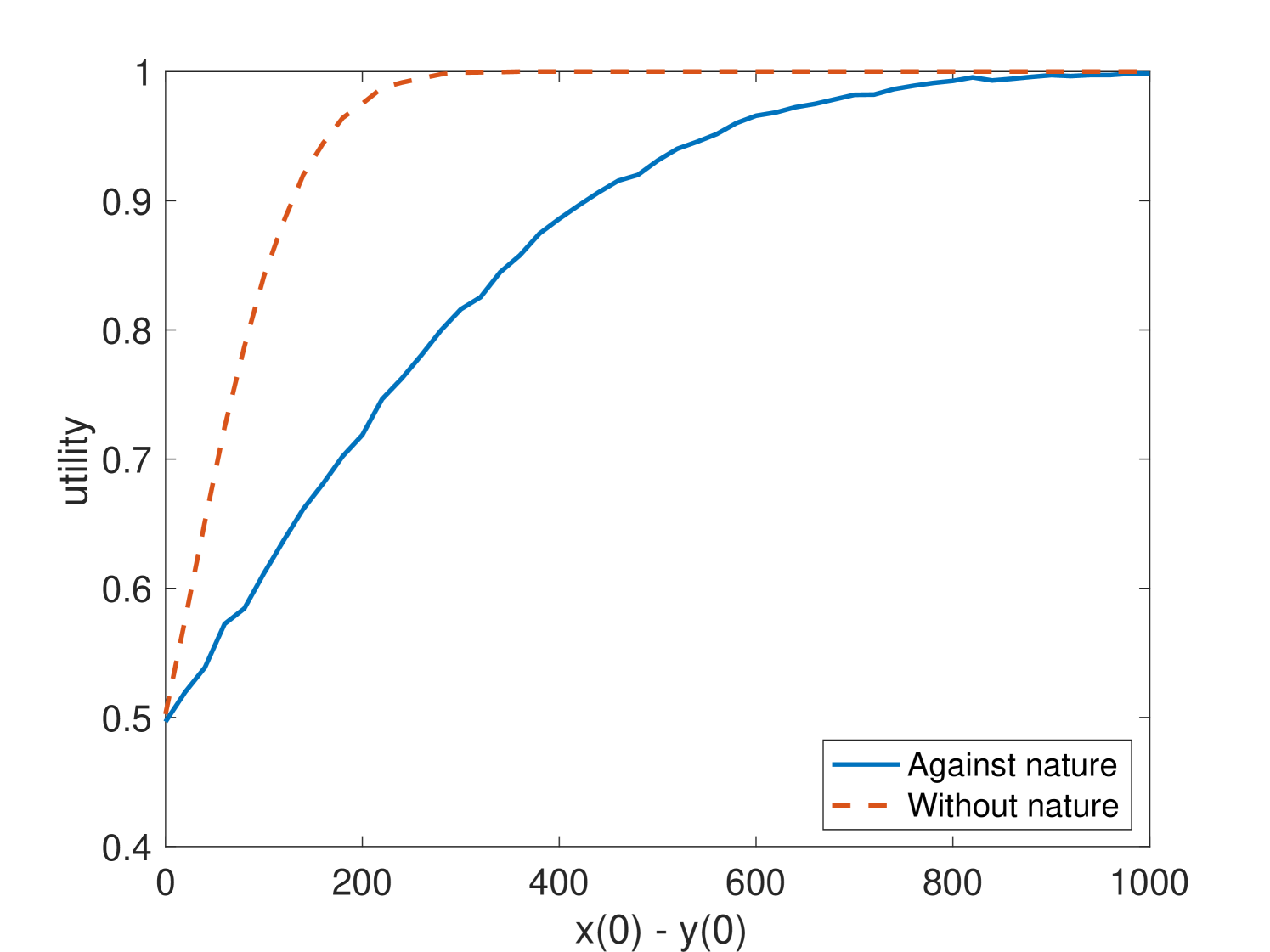
We first created models in MATLAB using SimBiology software tools for the catalyzed approximate majority CRN and the nature CRN described above. With initial populations and combined initial population , we varied the difference from 0 to 1,000 by intervals of 10. With these initial conditions, we ran in the presence of with . We ran 10,000 trials for each set of initial conditions. Each trial converged within time units, meaning that either or was 0. The design of the CRN guarantees that, once one population has taken over, no further reactions can occur.
As Fig. 1 shows, the random perturbations of and did reduce the success probability of the approximate majority algorithm for some values of . For example, when and , was successful in a vacuum but only successful in the presence of nature. However, even with the random perturbations, the success frequency in the presence of nature was always greater than of the success frequency in the absence of nature. This suggests that the CRN is at least -robust to in this game under arbitrary distributions of initial states with and .
5 Conclusion
Software engineering for molecular programming is a new research direction with open problems that can benefit from the attention of the software engineering research community. Many planned molecular systems will be deployed for use in vivo within a few years, and certification for safety-critical scenarios such as biosensors and drug delivery devices will require improved evidence of robustness. Molecular program developers similarly will be called upon to prevent system design vulnerabilities to malicious adversaries. Software engineering has an essential role to play in what scientists are already labeling as the century of life sciences [12].
The preliminary work described in this paper uses a game-theoretic approach to (1) formulate the robustness of a molecular program’s CRN model in terms of a game against nature and (2) provide a method to quantitatively evaluate its robustness. The example we present concerns random perturbations of the program’s clock speed by nature; however, the approach is general enough to also enable evaluation of security against an adversarial molecular program who maliciously perturbs the relative clock speeds. Future work on this will entail challenging issues involving strategic equilibria [18]. Our approach provides a foundation from which to pursue improved development and deployment of verifiably robust programmed molecular systems. More broadly, robustness in the presence of probabilistic behavior also is required for many non-molecular programmed systems [25, 11, 3, 20], and the advances described here may enhance our understanding of how to design in and verify robustness for other asynchronous systems operating in stochastic environments.
Acknowledgments
This research was supported in part by National Science Foundation grant 1545028. We thank Jim Lathrop for tool assistance.
References
- [1] David F. Anderson and Thomas G. Kurtz, editors. Stochastic Analysis of Biochemical Systems, volume 1.2 of Stochastics in Biological Systems. Springer International Publishing, 2015.
- [2] Stefan Badelt, Seung Woo Shin, Robert F. Johnson, Qing Dong, Chris Thachuk, and Erik Winfree. A general-purpose CRN-to-DSD compiler with formal verification, optimization, and simulation capabilities. In Proceedings of the 23rd International Conference on DNA Computing and Molecular Programming, Lecture Notes in Computer Science, pages 232–248, 2017.
- [3] Radu Calinescu, Carlo Ghezzi, Marta Z. Kwiatkowska, and Raffaela Mirandola. Self-adaptive software needs quantitative verification at runtime. Communications of the ACM, 55(9):69–77, 2012.
- [4] Yuan-Jyue Chen, Neil Dalchau, Niranjan Srinivas, Andrew Phillips, Luca Cardelli, David Soloveichik, and Georg Seelig. Programmable chemical controllers made from DNA. Nature Nanotechnology, 8(10):755–762, 2013.
- [5] Anne Condon, Monir Hajiaghayi, David G. Kirkpatrick, and Ján Manuch. Simplifying analyses of chemical reaction networks for approximate majority. In DNA Computing and Molecular Programming - 23rd International Conference, DNA 23, Austin, TX, USA, September 24-28, 2017, Proceedings, pages 188–209, 2017.
- [6] Matthew Cook, David Soloveichik, Erik Winfree, and Jehoshua Bruck. Programmability of chemical reaction networks. In Anne Condon, David Harel, Joost N. Kok, Arto Salomaa, and Erik Winfree, editors, Algorithmic Bioprocesses, Natural Computing Series, pages 543–584. Springer, 2009.
- [7] Frits Dannenberg, Marta Kwiatkowska, Chris Thachuk, and Andrew J. Turberfield. DNA walker circuits: Computational potential, design, and verification. In Proceedings of the 19th International Conference on DNA Computing and Molecular Programming, volume 8141 of Lecture Notes in Computer Science, pages 31–45. Springer, 2013.
- [8] René David and Hassane Alla. Discrete, Continuous, and Hybrid Petri Nets. Springer, 2010.
- [9] Shawn M. Douglas, Ido Bachelet, and George M. Church. A logic-gated nanorobot for targeted transport of molecular payloads. Science, 335(6070):831–834, 2012.
- [10] Samuel J. Ellis, Titus H. Klinge, James I. Lathrop, Jack H. Lutz, Robyn R. Lutz, Andrew S. Miner, and Hugh D. Potter. Runtime fault detection in programmed molecular systems. ACM Transactions on Software Engineering and Methodology, to appear.
- [11] Antonio Filieri, Carlo Ghezzi, and Giordano Tamburrelli. Run-time efficient probabilistic model checking. In Proceedings of the 33rd International Conference on Software Engineering, pages 341–350. ACM, 2011.
- [12] Jasmin Fisher, David Harel, and Thomas A. Henzinger. Biology as reactivity. Communications of the ACM, 54(10):72–82, 2011.
- [13] Daniel T. Gillespie. Exact stochastic simulation of coupled chemical reactions. The Journal of Physical Chemistry, 81(25):2340–2361, 1977.
- [14] John Knight. Fundamentals of Dependable Computing for Software Engineers. CRC Press, 2012.
- [15] Marta Kwiatkowska and Chris Thachuk. Probabilistic model checking for biology. Software Systems Safety, 36:165–189, 2014.
- [16] Robyn Lutz, Jack Lutz, James Lathrop, Titus Klinge, Eric Henderson, Divita Mathur, and Dalia Abo Sheasha. Engineering and verifying requirements for programmable self-assembling nanomachines. In Proceedings of the 34th International Conference on Software Engineering, pages 1361–1364. IEEE, 2012.
- [17] John Milnor. Games against nature. Technical report, Rand Corporation, 1951.
- [18] Abraham Neyman. Continuous-time stochastic games. Games and Economic Behavior, 104:92–130, 2017.
- [19] Christos H. Papadimitriou. Games against nature. J. Comput. Syst. Sci., 31(2):288–301, 1985.
- [20] Esteban Pavese, Víctor Braberman, and Sebastián Uchitel. Less is more: Estimating probabilistic rewards over partial system explorations. ACM Transactions on Software Engineering and Methodology, 25(2):16:1–16:47, 2016.
- [21] Lulu Qian, Erik Winfree, and Jehoshua Bruck. Neural network computation with DNA strand displacement cascades. Nature, 475(7356):368–372, 2011.
- [22] Y.A. Rozanov. Probability Theory: A Concise Course. Dover Publications, 1969.
- [23] David Soloveichik, Georg Seelig, and Erik Winfree. DNA as a universal substrate for chemical kinetics. Proceedings of the National Academy of Sciences, 107(12):5393–5398, 2010.
- [24] Niranjan Srinivas, James Parkin, Georg Seelig, Erik Winfree, and David Soloveichik. Enzyme-free nucleic acid dynamical systems. Science, 358(6369), 2017.
- [25] Guoxin Su, Taolue Chen, Yuan Feng, and David S. Rosenblum. ProEva: runtime proactive performance evaluation based on continuous-time markov chains. In Proceedings of the 39th International Conference on Software Engineering, ICSE 2017, Buenos Aires, Argentina, May 20-28, 2017, pages 484–495, 2017.
- [26] Axel van Lamsweerde. Requirements Engineering - From System Goals to UML Models to Software Specifications. Wiley, 2009.
- [27] Daniel Wilhelm, Jehoshua Bruck, and Lulu Qian. Probabilistic switching circuits in DNA. Proceedings of the National Academy of Sciences, 2018.