Supervised and Self-supervised Pretraining based COVID-19 detection using acoustic breathing/cough/speech signals
Abstract
In this work, we propose a bi-directional long short-term memory (BiLSTM) network based COVID-19 detection method using breath/speech/cough signals. By using the acoustic signals to train the network, respectively, we can build individual models for three tasks, whose parameters are averaged to obtain an average model, which is then used as the initialization for the BiLSTM model training of each task. This initialization method can significantly improve the performance on the three tasks, which surpasses the official baseline results. Besides, we also utilize a public pre-trained model wav2vec2.0 and pre-train it using the official DiCOVA datasets. This wav2vec2.0 model is utilized to extract high-level features of the sound as the model input to replace conventional mel-frequency cepstral coefficients (MFCC) features. Experimental results reveal that using high-level features together with MFCC features can improve the performance. To further improve the performance, we also deploy some preprocessing techniques like silent segment removal, amplitude normalization and time-frequency mask. The proposed detection model is evaluated on the DiCOVA dataset and results show that our method achieves an area under curve (AUC) score of 88.44% on blind test in the fusion track.
Index Terms— COVID-19, binary classification, supervised pre-training, self-supervised pre-training, respiratory diagnosis.
1 Introduction
Since the outbreak of COVID-19, it quickly becomes pandemic all over the world, even at every conner. This unknown disease has brought a serious influence to all countries ranging from global health, economy, education, trade, cultural exchange, etc. Due to the fact that disease diagnosis is an important step for controlling the transmission, it is worthy studying how to detect the COVID-19 rapidly and efficiently. Nucleic acid test is a common and traditional COVID-19 detection method, but patients have to be tested in specific place like hospitals, and the test is comparatively expensive and time-consuming. With the development of artificial intelligence (AI) techniques in recent years, machine learning based methods have been frequently used in disease detection. Compared to conventional medical diagnosis approaches, the AI-based methods have an obvious superiority, as the detection can be performed fully online and the time and economic expenses are much lower. The obtained diagnostic result can be used as supplementary information for doctors to make a more accurate clinical discrimination.
It thus deserves to investigate whether AI techniques can also be utilized for diagnosing COVID-19, where only medical images [1] or sounds [2] of patients will be recorded. Recently, several AI-based techniques have been developed for the detection of COVID-19. For example, a ResNet50 based method was proposed in [3] based on the use of computed tomography (CT) images, which achieves an accuracy of 96.23%. Compared to the CT-based method, where the CT images are collected offline, it is more appealing if we can use sound signals (e.g., cough, speech, breath) to perform detection, as the latter can be recorded remotely and avoid the people gathering.
In principal, the detection of COVID-19 is a binary classification task (i.e., positive or negative), for which convectional machine learning methods can be leveraged. In order to make use of sound data for classification, feature extraction is required as a pre-processing step. Mel-frequency cepstral coefficients (MFCC) and mel-frequency spectrogram that can reflect the nonlinear perceptive characteristics of human hearings to frequency are commonly used as features for sound activity analysis [4, 5]. In addition, zero crossing rate (ZCR), kurtosis, log energy, spectral centroid, roll-off frequency can also be used [6, 7]. In [8], it was shown that the positive testee of COVID-19 have different acoustical parameters compared with the negative. In [9], Coppock et al extracted the spectrogram feature and used a ResNet based CNN to detect cough sounds, which achieves an area under curve (AUC) of 0.846. In [10], the support vector machine (SVM) was employed for the detection of COVID-19 in combination of voice signals and symptoms. In addition, other classifiers like long short-term memory (LSTM) [6], k-Nearest Neighbor (kNN) [11], Random Forest [12] and Light gradient boosting machine (LightGBM) [13] can also be utilized in line with classical machine learning approaches.
As currently the amount of DiCOVA data is still limited, we mainly aim at exploring whether additional pre-training can improve the detection performance in this work. For this, we explore a supervised pre-training method, an unsupervised pre-training method and ensemble the two methods using the official dataset. Specifically, for supervised pre-training, we utilize breath, cough and speech sounds to train models, respectively, then we can obtain models for these three different tasks. The model parameters are averaged to obtain an average model. We treat the average model as an initialization model, which is taken to initialize the diagnosing model on different tasks. Experimental results show that this initialization method can significantly improve the performance on three tasks, which surpasses the official baseline results. For the unsupervised pre-training method, we pre-train the public pre-trained wav2vec2.0 model [14] on the DiCOVA dataset [15] and use the pre-trained model as a high-level feature extractor. We utilize the pre-trained model to extract high-level features on three tasks, which are input into the diagnosing model to replace the classic MFCC feature. Experimental results reveal that the developed unsupervised pre-training method also outperforms the official baseline, but it is worse than the supervised pre-training method. It is worth mentioning that ensembling the two methods can obtain a better result in the same task, which shows that ensemble different levels of information can improve the detection performance. More importantly, by fusing the output probabilities, we obtain the best performance in the fusion task.
2 Methodology
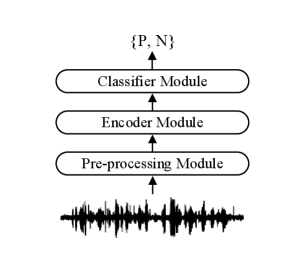
2.1 Model structure
The diagram of the proposed classification model that we utilize in each subtask is shown in Fig. 1, which mainly consists of a pre-processing module, an encoder module and a classifier module. In the encoder module, two bi-directional LSTM (BiLSTM) [16] layers was utilized as the encoder. Each BiLSTM layer contains 128 hidden units and the dropout rate is set to be 0.1. Two fully connected feed-forward layers are utilized in the classifier module, which consists of two linear transformations with a ReLU activation in between. As the detection of COVID-19 is a 0-1 classification problem, we use the binary classification loss function for training.
2.2 Supervised pre-training method
For the supervised pre-training, no extra data are utilized except the officially provided DiCOVA data. The total DiCOVA dataset consist of breathing, cough and speech labeled data. We obtain a breathing/cough/speech model using labeled breathing/cough/speech data through supervised pre-training, respectively. Specifically, the breathing model can be obtained using the labeled breathing data through supervised pre-training, and and can be built similarly as
| (1) | ||||
| (2) | ||||
| (3) |
Then, we average the parameters of the three models as the average model as
| (4) |
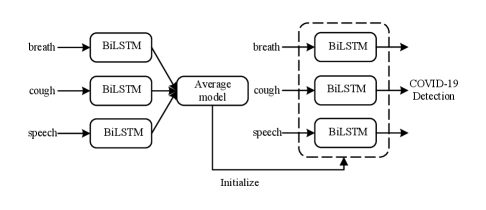
For each task, the average model is considered as an initialization model, and we re-train the breathing/cough/speech model in the corresponding task. The pre-training process is shown in Fig. 2. As we initialize the model using average model, the encoder and the classifier are easier to find the optimal solution.
2.3 Self-supervised pre-training method
As the amount of COVID-19 audio data in each sub-task is still limited, the traditional MFCC feature might be not sufficiently representative for classification tasks. Hence, we propose to utilize a high-level feature to replace the traditional MFCC feature. It is worth mentioning that there are many effective methods that were proposed to solve these low-resource tasks [17, 18, 19, 20, 21]. In the speech domain, The wav2vec2.0 [14] model is a representative self-supervised pre-training framework for learning speech representation. The public self-supervised pre-trained wav2vec2.0 [14] model is utilized in the stage of self-supervised pre-training. We pre-train the public pre-trained wav2vec2.0 model on the DiCOVA data and use the pre-trained model as a high-level feature extractor. For example, we use DiCOVA breathing data to pre-train the wav2vec2.0 model. After finishing pre-training, we utilize the pre-trained model to extract high-level breathing feature, which is then input into the diagnosing model to replace the MFCC feature.
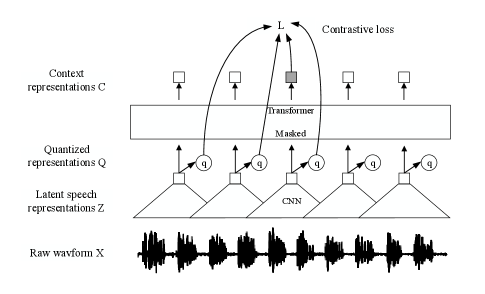
In order to guide the reader, we will briefly review the wav2vec 2.0 model in this section. The structure of wav2vec 2.0 model is shown in Fig. 3, including a CNN-based feature encoder and a transformer encoder . In detail, the input raw waveform is downsampled to the latent speech representation by the feature encoder. The transformer encoder then models the contextualized representation and extracts a high-level feature from the input . A quantization module discretizes the output of the feature encoder to as targets in the contrastive objective.
The quantization module first maps the latent speech representation to logits , given codebooks with entries.The Gumbel softmax operation [22] is then used to select discrete codebook entries in a fully differentiable way. For a given frame , we can therefore select one entry from each codebook and concatenate the resulting vectors and apply a linear transformation to obtain . The weighted loss function is thus given by
| (5) |
where
| (6) | ||||
| (7) | ||||
| (8) |
It is clear that the total loss function is the weighted summation of three terms , and parameterized by and . In (6), is the contrastive loss, which enables the model distinguishable between the true quantized latent speech representation and a set of quantized candidate representations . The quantized candidate representation contains and distractors, and the latter are uniformly sampled from other masked time steps of the same utterance. In (5), the diversity loss aims to increase the use of quantized codebook representation, and is an penalty over the outputs of the feature encoder. In (6), sim stands for the cosine similarity between two vectors and is a temperature. In (7), represents the probability of choosing the -th codebook entry for group across a batch of utterances, where is a temperature. In (8), stands for the average logits across utterances in a batch. More details on the wav2vec 2.0 model can be found in [14].
2.4 Model ensemble method
For the supervised pre-training method, the diagnosing model using MFCC feature as input, and for the self-supervised pre-training method, the diagnosing model using high-level features extracted by the pre-trained wav2vec2.0 model. As local features and high-level features contain different information, in order to combine the superiority of different features, we use model ensemble to improve the detection performance. Specifically, for the same task we utilize the proposed supervised and self-supervised methods to train two models and obtain and on the test set, respectively. Then, we can ensemble the scores obtained by the two models, which is given by
| (9) |
where is chosen by the user. The weighted summation is taken as the detection probability.
For the track-4 fusion task, we utilize the best breathing, cough and speech models to obtain an approporiate fusion model as
| (10) |
where the parameter need to satisfy . In principal, the breathing, cough and speech signals have a different importance in terms of the COVID-19 information, should be chosen in line with the signal qualities.
3 Performance evaluation
3.1 Datasets
The proposed model is evaluated on the DiCOVA-ICASSP 2022 challenge dataset [15], which is derived from the crowd-sourced Coswara dataset [23] and collected from volunteers with different health conditions. Volunteers were advised to record their sound in a quiet environment using a web-application. The audio streams are sampled at a sampling frequency of 44.1 kHz and in an FLAC format. The durations of the sound recordings range from about 1 second up to 29 seconds. The dataset consists of a total of 965 samples including 172 negative samples and 793 positive samples, where each sample includes cough, speech and breath sounds. Male and female patient samples are 723 and 242, respectively. The blind test set provided by the organizer includes 471 samples without labels.
3.2 Model configuration
In the pre-processing module, the amplitude of the raw waveform data is normalized between -1 to 1 through a normalization operator. As there are many silent segments in the speech/cough/breath sound signals, the speech activity detector (SAD) is applied to cut off these silent segments. The sound data is downsampled to 16 kHz. Forty dimensional MFCC and delta-delta coefficients are extracted with a widow of 25 msec audio samples and a hop of 10 msec. Due to the small size of the training data, we use SpecAugment [24] time-frequency mask to augment the data, as it was shown data augmentation is effective to improve the performance, particularly in low-resource cases. The time mask length is 20 and the frequency mask length is 50. In the encoder module, two BiLSTM [16] layers are utilized as the encoder. The BiLSTM layer dimension is 128 and the dropout rate is set to be 0.1. Two fully connected feed-forward layers are utilized in the classifier module, which consists of two linear transformations with a ReLU activation in between. The dimension of feed-forward layers is 256, which is finally mapped to 1-dimension for binary classification. As the detection of COVID-19 is a 0-1 classification problem, we use the binary classification loss function for training. We follow the baseline system given by the organizer to train the model for 5-fold cross-validation in each task and then decode on the official blind test set to obtain the final test results, separately. In the baseline system, The CNN model is also utilized as another baseline system in experiments.
For the supervised pre-training, different seeds are set to train different models. The pre-training model is trained using 2 RTX3090ti-24G GPUs with 50 epochs, and the total training time is about 1 hour. For the self-supervised pre-training, the wav2vec2.0 pre-training model is implemented using the fairseq toolkit [25]. The feature encoder contains seven blocks, where each block has 512 temporal convolution channels with strides (5, 2, 2, 2, 2, 2, 2) and kernel widths (10, 3, 3, 3, 3, 3, 2, 2). Thus, the interval between two sequential samples in the feature encoder output is around 20 ms and the receptive audio field is 25 ms. The models contain 12 transformer encoder blocks with a dimension of 512, a feed forward module with a dimension of 2048 and 8 attention heads. The pre-training process is optimized with Adam [26]. We set = 2 and = 320 for the quantization module and each entry with a size of 128. The temperature is set to be 0.1 and is annealed from 2 to 0.5 by a factor of 0.999995 over iterations. For the contrastive loss, and are set to be 0.1 and 10, respectively. We use = 100 distractors and the total number of pre-training epochs are 200. After self-supervised pre-training, the pre-trained model parameters are frozen. Then, the pre-trained model is utilized as a high-level feature extractor, which generates high-level features from raw waveforms.
| Method | Model | ROC-AUC score (%) | |
|---|---|---|---|
| Test | Validation | ||
| Track-1 breath | |||
| Official baseline | - | 84.50 | 77.63 |
| Baseline 1 | BiLSTM | 84.17 | 76.26 |
| Baseline 2 | CNN | 84.10 | 75.84 |
| Supervised pre-train | BiLSTM | 86.41 | 80.05 |
| Self-supervised pre-train | BiLSTM | 86.22 | 79.04 |
| Model ensemble | BiLSTM | 86.72 | 80.05 |
| Track-2 cough | |||
| Official baseline | - | 74.89 | 75.88 |
| Baseline 1 | BiLSTM | 75.04 | 76.18 |
| Baseline 2 | CNN | 73.70 | 76.06 |
| Supervised pre-train | BiLSTM | 76.05 | 78.92 |
| Self-supervised pre-train | BiLSTM | 75.55 | 78.56 |
| Model ensemble | BiLSTM | 76.36 | 78.92 |
| Track-3 speech | |||
| Official baseline | - | 84.26 | 82.24 |
| Baseline 1 | BiLSTM | 83.68 | 82.15 |
| Baseline 2 | CNN | 83.38 | 81.96 |
| Supervised pre-train | BiLSTM | 85.02 | 81.00 |
| Self-supervised pre-train | BiLSTM | 84.35 | 80.74 |
| Model ensemble | BiLSTM | 85.21 | 81.30 |
| Track-4 fusion | Fusion weight | ROC-AUC score (%) | |
|---|---|---|---|
| Test | Validation | ||
| Official baseline | - | 84.50 | 77.63 |
| Fusion1 | (1/3,1/3,1/3) | 87.01 | 82.56 |
| Fusion2 | (0.4,0.2,0.4) | 88.44 | 82.93 |
| Fusion3 | (0.5,0.1,0.4) | 88.44 | 82.71 |
3.3 Results
In experiments, we use 5-fold cross-validation to evaluate our supervised pre-training method, self-supervised pre-training method and model ensemble method. For comparison, we also test CNN, LSTM and official baseline model without pre-training. The official baseline comes from the website111https://competitions.codalab.org/competitions/34801#results. The obtained results are shown in Table 1 and Table 2. Experimental results show that the methods with pre-training achieve a higher AUC than the baseline. That is, pre-training can provide more information for classifiers, particularly in case the dataset is small-sized. Supervised pre-training models can achieve a great enhancement of performance compared with the baseline system. In breath/cough/speech task, supervised pre-training models increase the AUC from 84.50/74.89/84.26 to 86.41/76.05/85.02, respectively. Self-supervised pre-training methods obtain the AUC of 86.22/75.55/84.35 in the breath/cough/speech task. Despite achieving a smaller improvement in performance than supervised counterparts, self-supervised pre-training models still surpass the baseline in all tasks. Compared to the supervised and self-supervised methods, the proposed ensemble model can further improve the detection performance, which obtains the best performance for all tasks, and the corresponding AUCs are 86.72/76.36/85.21 in cough/speech/breath tasks, respectively. These results indicate that high-level feature pre-trained by wav2vec2.0 and low-level MFCC feature are complementary. For the fusion task, the final probability is calculated as a weighted summation over three best individual tasks. In case the cough/speech/breath weights are set to be 0.4/0.2/0.4, we achieve the highest AUC of 88.44 for the fusion task.
4 Conclusion and discussion
In this work, we presented an ensemble model based on BiLSTM for diagnosing COVID-19 using acoustic signals. Due to the small size of the training data, we used supervised pre-traing and self-supervised pre-training methods, and more importantly both achieved a better performance than the baseline. Using the ensemble model of supervised pre-traing and self-supervised pre-training, the AUC score was further improved, showing that the high-level feature pre-trained by wav2vec2.0 and low-level MFCC feature are complementary. The proposed model was evaluated on the DiCOVA challenge dataset and achieved an AUC score of 88.44% in the blind test set for the fusion task, which reaches the first place in tracks 3&4 of the DiCOVA-ICASSP 2022 contest.
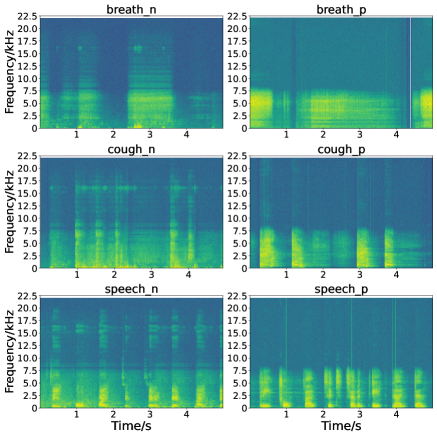
As it was shown by experiments that in case of using the same classifier the high-level representation obtained by the wav2vec 2.0 model achieves a better detection performance than the MFCC feature, it tells that feature extraction is a vital step for the COVID-19 detection. In order to more clearly see the difference in the features of positive and negative audio samples, we show the spectrograms of the positive and negative breathing/cough/speech audio samples in Fig. 4. It is clear that for the positive samples, the energy of breathing/cough/speech signals is almost concentrated on low-frequency bands, while for negative samples the energy is distributed over full-frequency bands, particularly for the negative cough and speech signals. It reveals that high-level features are more beneficial for diagnosing COVID-19 using acoustic signals. In the future, we will therefore investigate an effective combination of the wav2vec 2.0 based acoustic representation, spectrograms, MFCC as a more representative feature for this detection problem.
References
- [1] Md. Milon Islam, Fakhri Karray, Reda Alhajj, and Jia Zeng, “A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19),” IEEE Access, vol. 9, pp. 30551–30572, 2021.
- [2] Gauri Deshpande, Anton Batliner, and Björn W. Schuller, “AI-Based human audio processing for COVID-19: A comprehensive overview,” Pattern Recognition, vol. 122, pp. 108289, 2022.
- [3] Lucy Nwosu, Xiangfang Li, Lijun Qian, Seungchan Kim, and Xishuang Dong, “Semi-supervised learning for COVID-19 image classification via ResNet,” EAI Endorsed Transactions on Bioengineering and Bioinformatics, vol. 1, no. 3, 2021.
- [4] Vincent Karas and Björn W. Schuller, “Recognising covid-19 from coughing using ensembles of SVMs and LSTMs with handcrafted and deep audio features,” in Proc. Interspeech, 2021, pp. 911–915.
- [5] Kotra Venkata Sai Ritwik, Shareef Babu Kalluri, and Deepu Vijayasenan, “COVID-19 detection from spectral features on the DiCOVA dataset,” in Proc. Interspeech, 2021, pp. 936–940.
- [6] M. Pahar, M. Klopper, R. Warren, and T. Niesler, “COVID-19 cough classification using machine learning and global smartphone recordings,” Computers in Biology and Medicine, , no. 2, pp. 104572, 2021.
- [7] Chloe Brown, Jagmohan Chauhan, Andreas Grammenos, Jing Han, Apinan Hasthanasombat, Dimitris Spathis, Tong Xia, Pietro Cicuta, and Cecilia Mascolo, “Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data,” in Proc. of the ACM SIGKDD Conf. on Knowledge Discovery and Data Mining, 2020, pp. 3474–3484.
- [8] M. Asiaee, A. V. Azimi, S. S. Atashi, A. Keramatfar, and M. Nourbakhsh, “Voice quality evaluation in patients with COVID-19: An acoustic analysis,” Journal of Voice, 2020.
- [9] Harry Coppock, Alex Gaskell, Panagiotis Tzirakis, Alice Baird, Lyn Jones, and Björn Schuller, “End-to-end convolutional neural network enables COVID-19 detection from breath and cough audio: a pilot study,” BMJ Innovations, vol. 7, no. 2, pp. 356–362, 2021.
- [10] Jing Han, Chloë Brown, Jagmohan Chauhan, Andreas Grammenos, Apinan Hasthanasombat, Dimitris Spathis, Tong Xia, Pietro Cicuta, and Cecilia Mascolo, “Exploring automatic covid-19 diagnosis via voice and symptoms from crowdsourced data,” in Proc. IEEE Int. Conf. on Acoustics, Speech and Signal Processing (ICASSP), 2021, pp. 8328–8332.
- [11] Arup Anupam, N Jagan Mohan, Sudarsan Sahoo, and Sudipta Chakraborty, “Preliminary diagnosis of COVID-19 based on cough sounds using machine learning algorithms,” in IEEE Int. Conf. on Intelligent Computing and Control Systems, 2021, pp. 1391–1397.
- [12] Isabella Södergren, Maryam Nodeh, Prakash Chandra Chhipa, Konstantina Nikolaidou, and György Kovács, “Detecting COVID-19 from audio recording of coughs using Random Forests and Support Vector Machines,” in Proc. Interspeech, 2021, pp. 916–920.
- [13] Madhu Kamble, Jose Gonzalez Lopez, Teresa Grau, Juan Espín López, Lorenzo Cascioli, Yiqing Huang, Alejandro Gomez-Alanis, Jose Patino, Roberto Font, Antonio Peinado, Angel Gomez, Nicholas Evans, Maria Zuluaga, and Massimiliano Todisco, “PANACEA cough sound-based diagnosis of COVID-19 for the DiCOVA 2021 challenge,” in Proc. Interspeech, 2021, pp. 906–910.
- [14] Alexei Baevski, Yuhao Zhou, Abdelrahman Mohamed, and Michael Auli, “wav2vec 2.0: A framework for self-supervised learning of speech representations,” Advances in Neural Information Processing Systems, vol. 33, 2020.
- [15] Neeraj Kumar Sharma, Srikanth Raj Chetupalli, Debarpan Bhattacharya, Debottam Dutta, Pravin Mote, and Sriram Ganapathy, “The Second DiCOVA Challenge: Dataset, task, and baseline system for COVID-19 diagnosis using acoustics,” https://dicovachallenge.github.io/docs/Second_DiCOVA2_baselineSystem_doc.pdf.
- [16] Sepp Hochreiter and Jürgen Schmidhuber, “Long short-term memory,” Neural Computation, vol. 9, no. 8, pp. 1735–1780, 1997.
- [17] A. van den Oord, Y. Li, and O. Vinyals, “Representation learning with contrastive predictive coding,” arXiv preprint arXiv:1807.03748, 2018.
- [18] J. Chorowski, R. J. Weiss, S. Bengio, and A. van den Oord, “Unsupervised speech representation learning using wavenet autoencoders,” IEEE/ACM Tran. Audio, Speech, Language Process., vol. 27, no. 12, pp. 2041–2053, 2019.
- [19] Yu-An Chung, Wei-Ning Hsu, Hao Tang, and James Glass, “An unsupervised autoregressive model for speech representation learning,” in Proc. Interspeech, 2019, pp. 146–150.
- [20] Steffen Schneider, Alexei Baevski, Ronan Collobert, and Michael Auli, “Wav2vec: unsupervised pre-training for speech recognition,” in Proc. Interspeech, 2019, pp. 3465–3469.
- [21] Alexei Baevski, Steffen Schneider, and Michael Auli, “vq-wav2vec: Self-supervised learning of discrete speech representations,” arXiv preprint arXiv:1910.05453, 2019.
- [22] Eric Jang, Shixiang Gu, and Ben Poole, “Categorical reparameterization with gumbel-softmax,” arXiv preprint arXiv:1611.01144, 2016.
- [23] N Sharma, P Krishnan, R Kumar, S Ramoji, SR Chetupalli, R Nirmala, P Kumar Ghosh, and Ganapathy, “Coswara - A database of breathing, cough, and voice sounds for COVID-19 diagnosis,” in Proc. Interspeech, 2020, pp. 4811–4815.
- [24] Daniel Park, William Chan, Yu Zhang, Chung-Cheng Chiu, Barret Zoph, Ekin Cubuk, and Quoc Le, “SpecAugment: A simple data augmentation method for automatic speech recognition,” in Proc. Interspeech, 2019, pp. 2613–2617.
- [25] Myle Ott, Sergey Edunov, Alexei Baevski, Angela Fan, Sam Gross, Nathan Ng, David Grangier, and Michael Auli, “Fairseq: A fast, extensible toolkit for sequence modeling,” in Proc. Conf. the North American Chapter of the Association for Computational Linguistics (Demonstrations), 2019, pp. 48–53.
- [26] Diederik P Kingma and Jimmy Ba, “Adam: A method for stochastic optimization,” arXiv preprint arXiv:1412.6980, 2014.