Task-adaptive Pre-training of Language Models with Word Embedding Regularization
Abstract
Pre-trained language models (PTLMs) acquire domain-independent linguistic knowledge through pre-training with massive textual resources. Additional pre-training is effective in adapting PTLMs to domains that are not well covered by the pre-training corpora. Here, we focus on the static word embeddings of PTLMs for domain adaptation to teach PTLMs domain-specific meanings of words. We propose a novel fine-tuning process: task-adaptive pre-training with word embedding regularization (TAPTER). TAPTER runs additional pre-training by making the static word embeddings of a PTLM close to the word embeddings obtained in the target domain with fastText. TAPTER requires no additional corpus except for the training data of the downstream task. We confirmed that TAPTER improves the performance of the standard fine-tuning and the task-adaptive pre-training on BioASQ (question answering in the biomedical domain) and on SQuAD (the Wikipedia domain) when their pre-training corpora were not dominated by in-domain data.
1 Introduction
Pre-trained language models (PTLMs) trained with massive textual and computational resources have achieved high performance in natural language processing tasks Devlin et al. (2019). Additional pre-training often is used to tackle domain discrepancies between the downstream task and the pre-training corpora. Additional pre-training with a large corpus in the domain of the downstream task, such as BioBERT Lee et al. (2020), improves the performance on the task Alsentzer et al. (2019); Beltagy et al. (2019); Chalkidis et al. (2020). However, this approach requires large corpora in the target domain and entails a high computational cost.
Gururangan et al. (2020) proposed task-adaptive pre-training (TAPT), which is additional pre-training using only the training data of the downstream task. TAPT can be regarded as a new fine-tuning process in which the standard fine-tuning is preceded by low-cost additional pre-training.
In this study, we focus on the static word embeddings of PTLMs (i.e., non-contextualized 0-th layer representations) for domain adaptation. Our method is designed to teach PTLMs the domain-specific meanings of the words as static word embeddings. We are motivated by the observation that the middle BERT layers capture the syntactic information Hewitt and Manning (2019); Jawahar et al. (2019); Liu et al. (2019a). We consider that we can adapt the models without harming the domain-independent linguistic knowledge contained in higher layers by learning the static word embeddings directly.
We propose a novel fine-tuning process called task-adaptive pre-training with word embedding regularization (TAPTER). First, TAPTER obtains word embeddings in the target domain by adapting a pre-trained fastText model Bojanowski et al. (2017) to the target domain using the training data of the downstream task. Next, TAPTER runs the task-adaptive pre-training by making the static word embeddings of the PTLM close to the word embeddings obtained with the fastText model. Finally, TAPTER runs the standard fine-tuning process.
We found that TAPTER achieves higher scores than the standard fine-tuning and TAPT on question answering tasks in the biomedical domain, BioASQ Tsatsaronis et al. (2015), and Wikipedia domain, SQuAD1.1 Rajpurkar et al. (2016). Our key findings are: (i) Word embedding regularization in task-adaptive pre-training enhances domain adaptation when the initial pre-training corpora do not contain a high proportion of in-domain data. (ii) The word embeddings of fastText, which uses a shallow neural network, can be adapted to the target domains more easily than the static word embeddings of PTLMs.
2 Preliminaries
2.1 Pre-trained Language Models
We focus on the static word embeddings of PTLMs. Let be the vocabulary. We input a token sequence to the model, where is the length of the sequence. The embedding layer of the model has a word embedding matrix as trainable parameters, where is the embedding dimension. The word embedding of the -th token is .
2.2 fastText
fastText is a word embedding method using subword information Bojanowski et al. (2017). The skipgram model Mikolov et al. (2013) of fastText learns word embeddings by predicting the surrounding words from a word , where is the set of the indices within a given window size. Specifically at position , we use the surrounding words as positive examples and randomly sample negative words from the vocabulary . The loss function is
That is, the model learns to score higher for positive examples and lower for negative examples.
fastText uses subword information to model the score function . Let be the set of substrings of the word . The score of the input word and the output word is
Here, consists of the word embeddings of the input layer and consists of the word embeddings of the output layer. is the embedding dimension, and is an arbitrary large number that determines the actual vocabulary size of the subwords. In the implementation of fastText, the model does not restrict the vocabulary size by hashing a subword into an index less than . The model has limits on the minimum and maximum lengths of subwords.
At inference time, the embedding of a word is . Bojanowski et al. (2017) reported that fastText learns word similarity with less training data than other methods do by utilizing the subword information.
2.3 Related Work
Static word embeddings in PTLMs have attracted attention in the areas of domain adaptation and cross-lingual transfer learning. Artetxe et al. (2020) proposed to replace word embeddings in the PTLMs trained in the source or target languages. Poerner et al. (2020) proposed a vocabulary expansion using Word2Vec Mikolov et al. (2013) trained in the target domain for domain adaptation on a CPU. However, our preliminary experiments showed that simple replacement or vocabulary expansion harms performance in our setting because of the limited amount of data. Unlike the previous studies, the proposed method requires no additional corpus by incorporating regularization of the word embeddings in the additional pre-training framework with the training data of the downstream task.
3 Proposed Method
The proposed method consists of three stages.
Additional Training of fastText
First, we train a fastText model using the training data of the downstream task, where the model is initialized with publicly available fastText embeddings111https://fasttext.cc/.
Our method introduces the embeddings of the PTLM vocabulary inferred by using the fastText model as the word embeddings in the target domain. Unlike other word embedding methods such as GloVe Pennington et al. (2014), fastText retains subword information. Therefore, we can obtain the embeddings of the PTLM vocabulary containing subword units222We cannot obtain the embeddings of subwords shorter than the minimum length configured in fastText. The proposed method ignores such subwords in the additional training of the PTLMs.. The additional training of fastText runs much faster than the additional training of the PTLMs. Note that TAPTER does not make any changes to the original vocabulary of the PTLMs.
Additional Pre-training of PTLMs
Second, we train the entire PTLM using the training data of the downstream task. We input a token sequence to the model. We train the model with the loss function of language modeling with -norm regularization on the difference between the word embeddings. That is, the loss function is
| (1) |
where is the set of the target tokens of the regularization. The target tokens exclude stop words and subwords shorter than the minimum length configured in fastText.
The function maps a -dimensional embedding to a -dimensional embedding:
LN denotes layer normalization Ba et al. (2016). are trainable parameters. The loss function of Eq. (1) is designed to alleviate the catastrophic forgetting problem with the first term and to adapt the word embeddings to the target domain with the second term. Miceli Barone et al. (2017); März et al. (2019) proposed -norm regularization for domain adaptation, but they did not incorporate it in the additional pre-training framework.
Fine-Tuning
Finally, we run the standard fine-tuning process Devlin et al. (2019) without any regularization.
4 Evaluation
4.1 Dataset
We evaluated the proposed method on two question answering datasets. Table 1 shows the statistics. The experimental setup is shown in Appendix A.
| Training | Evaluation | Corpus Size | |
|---|---|---|---|
| SQuAD1.1 | 87,599 | 10,570 | 2.62M |
| BioASQ5 | 4,950 | 150 | 1.38M |
SQuAD
SQuAD1.1 is a task to answer a question with information from a textual source Rajpurkar et al. (2016). The dataset provides pairs of a question and a related passage from Wikipedia as the textual source. The input of the model is a token sequence that is a concatenation of the question and the passage. The ground-truth answer is a span in the passage. The output of the model consists of the indices of the answer start and end positions. The indices are calculated from the two-dimensional linear layer on top of the PTLM. The official evaluation metrics are exact matching (EM) and partial matching (F1).
BioASQ
BioASQ5 is a question answering dataset in the biomedical domain Tsatsaronis et al. (2015). Following Lee et al. (2020), we used the factoid questions pre-processed into SQuAD format. We used three official evaluation metrics. Strict accuracy (SACC) is the rate at which the top-1 prediction is correct. Lenient accuracy (LACC) is the rate at which the top-5 predictions include the ground-truth answer. Mean reciprocal rank (MRR) is the average of the reciprocal of the rank of the ground-truth answer. We trained the models with ten random seeds and report the average performance. In the fine-tuning stage, as in Wiese et al. (2017), we first trained the model with SQuAD and then trained it with BioASQ.
4.2 Compared Models
We used three PTLMs, BERT-base-cased Devlin et al. (2019), BioBERT Lee et al. (2020), and RoBERTa-base Liu et al. (2019b). BERT-base-cased was pre-trained with English Wikipedia (2.5B words) and BookCorpus (800M words) Zhu et al. (2015). BioBERT was initialized with BERT-base-cased and pre-trained with PubMed abstracts and PMC articles (18B words). RoBERTa-base was pre-trained with 160GB corpora including news and Web articles as well as Wikipedia and BookCorpus (used to train BERT). We compared three fine-tuning methods: standard fine-tuning, TAPT, and TAPTER.
4.3 Results and Discussion
Is TAPTER effective at adaptation to the biomedical domain?
Table 2 shows the results in BioASQ. We evaluated the performance of the domain adaptation with BERT-base-cased because the model does not use a biomedical corpus in the original pre-training.
TAPTER improved BERT’s performance by 3.05/0.27/2.01 points (SACC/LACC/MRR) over the simple fine-tuning. As well, TAPTER statistically significantly outperformed TAPT in the SACC (top-1 accuracy) and MRR metrics. We consider that the regularization of the word embeddings improves the adaptation of the PTLM.
Appendix B shows the word embeddings of the models with principal component analysis. The scatter plots show that the word embeddings of BERT-base-cased and TAPT resemble each other, though TAPTER and BioBERT have dissimilar word embeddings distributions to that of BERT-base-cased. This indicates that the additional pre-training of language modeling alone does not adapt the static word embeddings to the biomedical domain unlike TAPTER.
| SACC | LACC | MRR | |
|---|---|---|---|
| BERT-base-cased | 37.88 | 54.00 | 43.87 |
| +TAPT | 39.47 | 54.27 | 44.69 |
| +TAPTER | 40.93** | 54.27 | 45.88** |
| BioBERT | 43.53 | 59.67 | 49.81 |
| +TAPT | 45.67 | 57.87 | 50.46 |
| +TAPTER | 44.60 | 58.33 | 50.02 |
Is additional pre-training effective with the model pre-trained in the target domain?
The additional pre-training from BioBERT did not improve the overall performance, although some of the scores slightly increased. There was no significant difference between TAPTER and TAPT in each metric (). We consider that BioBERT has already learned the knowledge in the biomedical domain because it was pre-trained with a massive biomedical text. Therefore, the additional pre-training had little effect on performance.
Is TAPTER effective in the general domain?
| EM | F1 | |
|---|---|---|
| BERT-base-cased | 79.12 | 87.55 |
| +TAPT | 78.42 | 87.12 |
| +TAPTER | 78.68 | 87.19 |
| RoBERTa-base | 82.76 | 90.40 |
| +TAPT | 83.01 | 90.45 |
| +TAPTER | 83.55 | 90.86 |
Table 3 shows the results for SQuAD. In the experiments with BERT neither of the additional pre-training methods improved performance. On the other hand, in the experiments with RoBERTa, TAPTER improved performance by 0.79/0.46 points (EM/F1). This was the best performance among the compared models on SQuAD.
We consider that TAPTER and TAPT improve performance when the corpora of the original pre-training were not dominated by in-domain data. A large part of the pre-training corpora of BERT is Wikipedia. Therefore, the additional pre-training was not effective. However, the pre-training corpora of RoBERTa cover broader topics. Although the corpora include Wikipedia, the additional pre-training can adapt the model to the Wikipedia domain.
It is known that the performance of PTLMs tends to improve as the amount of pre-training corpora increases Baevski et al. (2019); Lan et al. (2019). Our results show that TAPTER can improve the performance of PTLMs that were pre-trained with very large corpora even if the domain of the downstream task is included in the pre-training corpora.
How well does TAPTER learn the language modeling and the word embeddings?
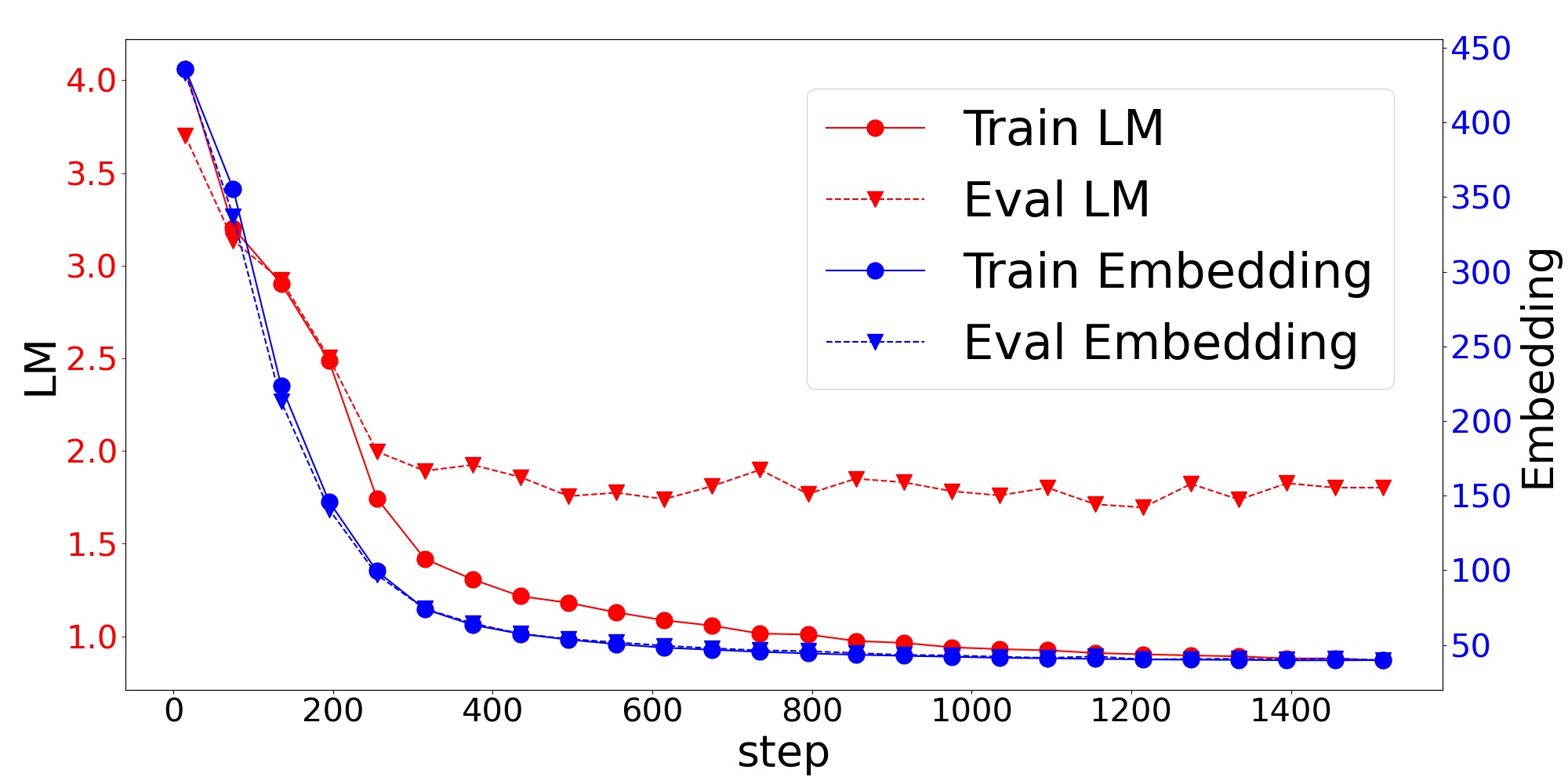
Figure 1 shows the learning curve of the additional pre-training in BioASQ from BERT. We can see that the second term in Eq. (1) representing the word embeddings decreased more sharply than the first term in Eq. (1) representing the language modeling. Since the BERT model is huge and complicated, we must learn the model slowly with a small learning rate over a large number of steps. However, the regularization term decreases quickly without corrupting the model. This is one of the advantages of TAPTER in low-resource settings.
In addition, the decrease in the first term on the development data stopped on the way. However, the word embeddings were trained with less discrepancy between the training and development data. We consider that the training data of BioASQ is too small to represent the distribution of the text in the biomedical domain. Since MLM takes a document-level sequence X as input, the search space of the true distribution is huge, and MLM is a very difficult task to train with limited training data. On the other hand, the regularization term depends on the word-level distribution . Therefore, the model can decrease the regularization term on the evaluation data even in low-resource settings without overfitting.
5 Conclusion
We proposed a new fine-tuning process including additional pre-training with word embedding regularization. TAPTER learns the meanings of words in the target domain by making the static word embeddings of the PTLM close to the word embeddings obtained in the target domain with fastText. TAPTER improves the performance of BERT in the biomedical domain. Moreover, it improves the performance of RoBERTa even in the Wikipedia domain although the original pre-training corpora of RoBERTa contain Wikipedia.
References
- Alsentzer et al. (2019) Emily Alsentzer, John Murphy, William Boag, Wei-Hung Weng, Di Jindi, Tristan Naumann, and Matthew McDermott. 2019. Publicly available clinical BERT embeddings. In Proceedings of the 2nd Clinical NLP@ACL, pages 72–78.
- Artetxe et al. (2020) Mikel Artetxe, Sebastian Ruder, and Dani Yogatama. 2020. On the cross-lingual transferability of monolingual representations. In ACL, pages 4623–4637.
- Ba et al. (2016) Jimmy Lei Ba, Jamie Ryan Kiros, and Geoffrey E Hinton. 2016. Layer normalization. arXiv preprint arXiv:1607.06450.
- Baevski et al. (2019) Alexei Baevski, Sergey Edunov, Yinhan Liu, Luke Zettlemoyer, and Michael Auli. 2019. Cloze-driven pretraining of self-attention networks. In EMNLP-IJCNLP, pages 5360–5369.
- Beltagy et al. (2019) Iz Beltagy, Kyle Lo, and Arman Cohan. 2019. SciBERT: A pretrained language model for scientific text. In EMNLP-IJCNLP, pages 3615–3620.
- Bird et al. (2009) Steven Bird, Ewan Klein, and Edward Loper. 2009. Natural Language Processing with Python. O’Reilly Media, Inc.
- Bojanowski et al. (2017) Piotr Bojanowski, Edouard Grave, Armand Joulin, and Tomas Mikolov. 2017. Enriching word vectors with subword information. TACL, 5:135–146.
- Chalkidis et al. (2020) Ilias Chalkidis, Manos Fergadiotis, Prodromos Malakasiotis, Nikolaos Aletras, and Ion Androutsopoulos. 2020. LEGAL-BERT: The muppets straight out of law school. In EMNLP Findings, pages 2898–2904, Online.
- Devlin et al. (2019) Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2019. BERT: Pre-training of deep bidirectional transformers for language understanding. NAACL-HLT, pages 4171–4186.
- Gururangan et al. (2020) Suchin Gururangan, Ana Marasović, Swabha Swayamdipta, Kyle Lo, Iz Beltagy, Doug Downey, and Noah A. Smith. 2020. Don’t stop pretraining: Adapt language models to domains and tasks. In ACL, pages 8342–8360.
- Hewitt and Manning (2019) John Hewitt and Christopher D. Manning. 2019. A structural probe for finding syntax in word representations. In NAACL-HLT, pages 4129–4138.
- Honnibal et al. (2020) Matthew Honnibal, Ines Montani, Sofie Van Landeghem, and Adriane Boyd. 2020. spaCy: Industrial-strength Natural Language Processing in Python.
- Jawahar et al. (2019) Ganesh Jawahar, Benoît Sagot, and Djamé Seddah. 2019. What does BERT learn about the structure of language? In ACL, pages 3651–3657, Florence, Italy.
- Kingma and Ba (2014) Diederik P. Kingma and Jimmy Ba. 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
- Lan et al. (2019) Zhenzhong Lan, Mingda Chen, Sebastian Goodman, Kevin Gimpel, Piyush Sharma, and Radu Soricut. 2019. Albert: A lite bert for self-supervised learning of language representations. In ICLR.
- Lee et al. (2020) Jinhyuk Lee, Wonjin Yoon, Sungdong Kim, Donghyeon Kim, Sunkyu Kim, Chan Ho So, and Jaewoo Kang. 2020. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4):1234–1240.
- Liu et al. (2019a) Nelson F. Liu, Matt Gardner, Yonatan Belinkov, Matthew E. Peters, and Noah A. Smith. 2019a. Linguistic knowledge and transferability of contextual representations. In NAACL-HLT, pages 1073–1094.
- Liu et al. (2019b) Yinhan Liu, Myle Ott, Naman Goyal, Jingfei Du, Mandar Joshi, Danqi Chen, Omer Levy, Mike Lewis, Luke Zettlemoyer, and Veselin Stoyanov. 2019b. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692.
- März et al. (2019) Luisa März, Dietrich Trautmann, and Benjamin Roth. 2019. Domain adaptation for part-of-speech tagging of noisy user-generated text. In NAACL-HLT, pages 3415–3420.
- Miceli Barone et al. (2017) Antonio Valerio Miceli Barone, Barry Haddow, Ulrich Germann, and Rico Sennrich. 2017. Regularization techniques for fine-tuning in neural machine translation. In EMNLP, pages 1489–1494.
- Mikolov et al. (2013) Tomas Mikolov, Ilya Sutskever, Kai Chen, Greg S Corrado, and Jeff Dean. 2013. Distributed representations of words and phrases and their compositionality. NIPS, 26:3111–3119.
- Paszke et al. (2019) Adam Paszke, Sam Gross, Francisco Massa, Adam Lerer, James Bradbury, Gregory Chanan, Trevor Killeen, Zeming Lin, Natalia Gimelshein, Luca Antiga, Alban Desmaison, Andreas Kopf, Edward Yang, Zachary DeVito, Martin Raison, Alykhan Tejani, Sasank Chilamkurthy, Benoit Steiner, Lu Fang, Junjie Bai, and Soumith Chintala. 2019. Pytorch: An imperative style, high-performance deep learning library. In NIPS, pages 8024–8035.
- Pennington et al. (2014) Jeffrey Pennington, Richard Socher, and Christopher D. Manning. 2014. GloVe: Global vectors for word representation. In EMNLP, pages 1532–1543.
- Poerner et al. (2020) Nina Poerner, Ulli Waltinger, and Hinrich Schütze. 2020. Inexpensive domain adaptation of pretrained language models: Case studies on biomedical NER and covid-19 QA. In EMNLP Findings, pages 1482–1490.
- Raffel et al. (2020) Colin Raffel, Noam Shazeer, Adam Roberts, Katherine Lee, Sharan Narang, Michael Matena, Yanqi Zhou, Wei Li, and Peter J Liu. 2020. Exploring the limits of transfer learning with a unified text-to-text transformer. JMLR, 21:1–67.
- Rajpurkar et al. (2016) Pranav Rajpurkar, Jian Zhang, Konstantin Lopyrev, and Percy Liang. 2016. SQuAD: 100,000+ questions for machine comprehension of text. In EMNLP, pages 2383–2392.
- Sennrich et al. (2016) Rico Sennrich, Barry Haddow, and Alexandra Birch. 2016. Neural machine translation of rare words with subword units. In ACL, pages 1715–1725.
- Shoeybi et al. (2019) Mohammad Shoeybi, Mostofa Patwary, Raul Puri, Patrick LeGresley, Jared Casper, and Bryan Catanzaro. 2019. Megatron-lm: Training multi-billion parameter language models using gpu model parallelism. arXiv preprint arXiv:1909.08053.
- Tsatsaronis et al. (2015) George Tsatsaronis, Georgios Balikas, Prodromos Malakasiotis, Ioannis Partalas, Matthias Zschunke, Michael R Alvers, Dirk Weissenborn, Anastasia Krithara, Sergios Petridis, Dimitris Polychronopoulos, Yannis Almirantis, John Pavlopoulos, Nicolas Baskiotis, Patrick Gallinari, Thierry Artieres, Axel Ngonga, Norman Heino, Eric Gaussier, Liliana Barrio-Alvers, Michael Schroeder, Ion Androutsopoulos, and Georgios Paliouras. 2015. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinformatics, 16:138.
- Wiese et al. (2017) Georg Wiese, Dirk Weissenborn, and Mariana Neves. 2017. Neural domain adaptation for biomedical question answering. In CoNLL 2017, pages 281–289.
- Wolf et al. (2020) Thomas Wolf, Lysandre Debut, Victor Sanh, Julien Chaumond, Clement Delangue, Anthony Moi, Pierric Cistac, Tim Rault, Rémi Louf, Morgan Funtowicz, Joe Davison, Sam Shleifer, Patrick von Platen, Clara Ma, Yacine Jernite, Julien Plu, Canwen Xu, Teven Le Scao, Sylvain Gugger, Mariama Drame, Quentin Lhoest, and Alexander M. Rush. 2020. Transformers: State-of-the-art natural language processing. In EMNLP: System Demonstrations, pages 38–45.
- Wu et al. (2016) Yonghui Wu, Mike Schuster, Zhifeng Chen, Quoc V Le, Mohammad Norouzi, Wolfgang Macherey, Maxim Krikun, Yuan Cao, Qin Gao, Klaus Macherey, et al. 2016. Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv preprint arXiv:1609.08144.
- Zhu et al. (2015) Yukun Zhu, Ryan Kiros, Rich Zemel, Ruslan Salakhutdinov, Raquel Urtasun, Antonio Torralba, and Sanja Fidler. 2015. Aligning books and movies: Towards story-like visual explanations by watching movies and reading books. In ICCV, pages 19–27.
Appendix A Experimental Setup
We trained the models on one NVIDIA GeForce GTX 1080Ti (11GB). The hyperparameter settings are in Table 4 and Table 5. The optimization algorithm was Adam Kingma and Ba (2014). We used PyTorch Paszke et al. (2019) and Transformers Wolf et al. (2020). Stop words were implemented by NLTK Bird et al. (2009). The word-level tokenizer was spaCy Honnibal et al. (2020). For the target tokens of the regularization , we randomly selected 50 % tokens in the input excluding stop words and subwords shorter than the minimum length configured in fastText. Following the default setting, the minimum length of the subwords in fastText was set to three. The maximum length was six. In BioASQ, we lowercased the corpora in the additional training of fastText and in the additional pre-training because only a limited number of words containing capital characters appear.
Note that the computational time of our additional pre-training was about seven hours on one NVIDIA GeForce GTX 1080Ti (11GB) GPU, while that of BioBERT was more than ten days on eight NVIDIA V100 (32GB) GPUs.
| Pre-Training | Fine-Tuning | |
|---|---|---|
| batch size | 256 | 128 |
| epochs | 100 | 5 / 2 / 10 |
| max seq. length | 512 | 384 |
| max query length | – | 64 |
| learning rate | 5e-5 | |
| warmup proportion | 0.06 | |
| weight decay | 0.01 | |
| SQuAD | BioASQ | |
|---|---|---|
| min count | 5 | 2 |
| epochs | 5 | 10 |
| dim | 300 | |
Appendix B Visualization of Word Embeddings
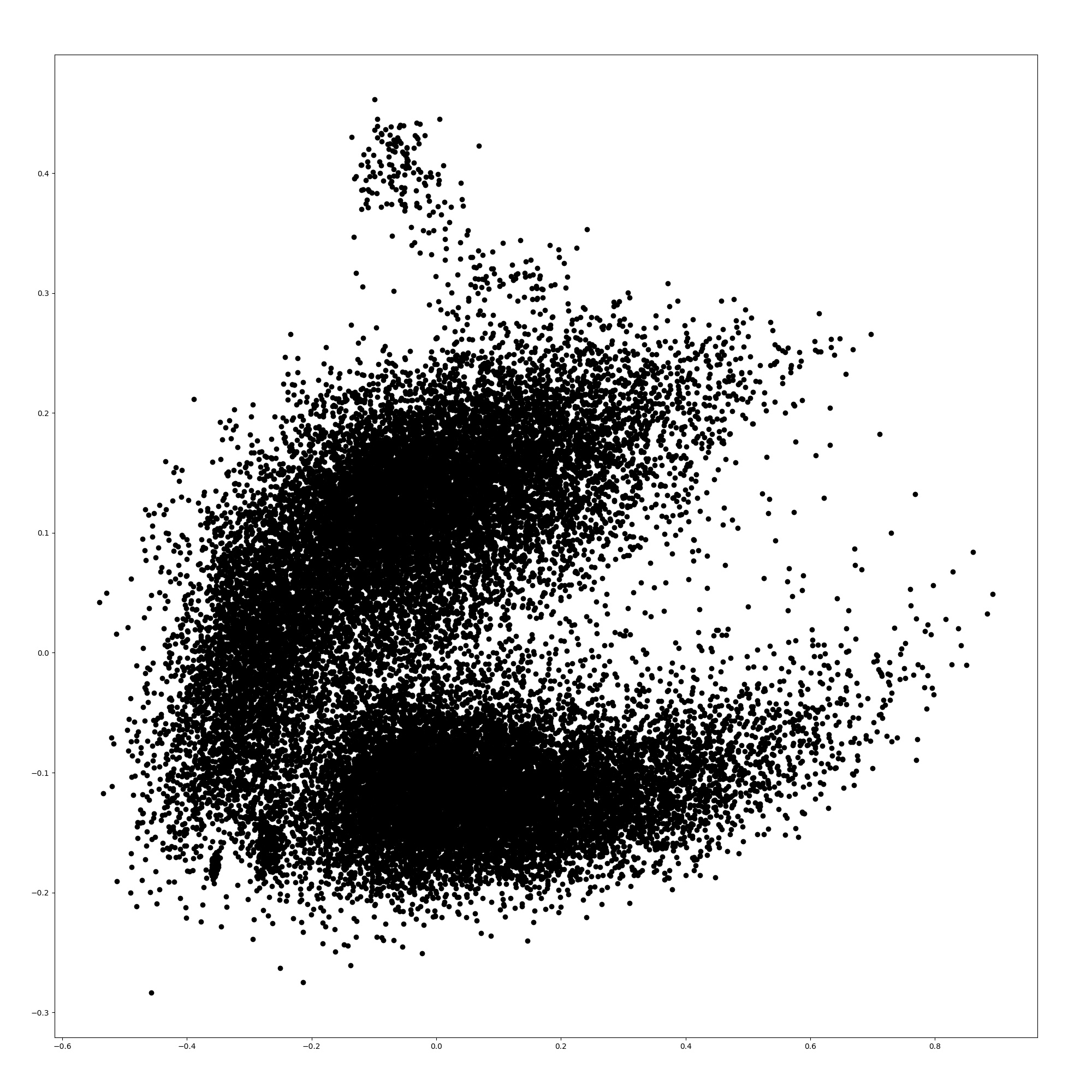
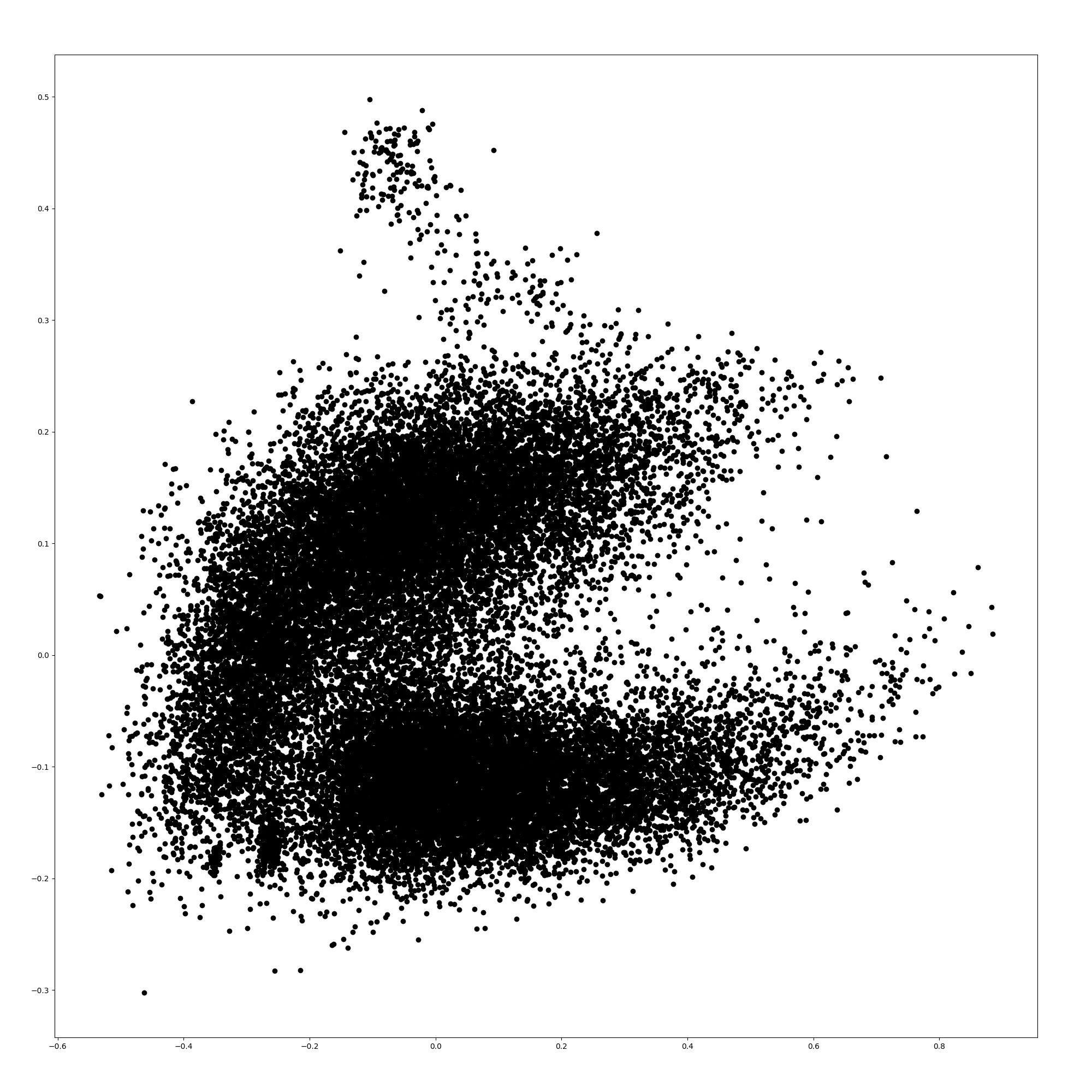
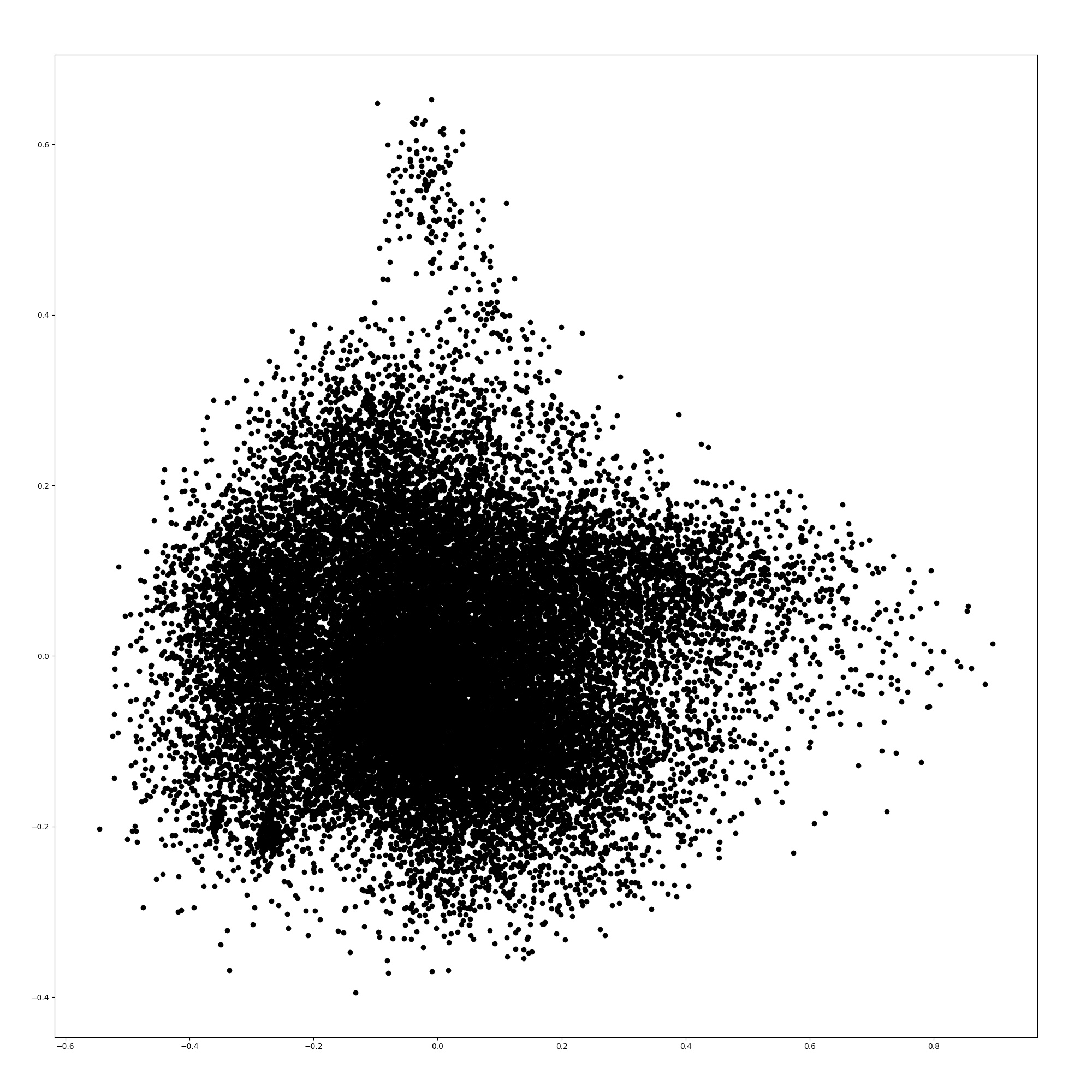
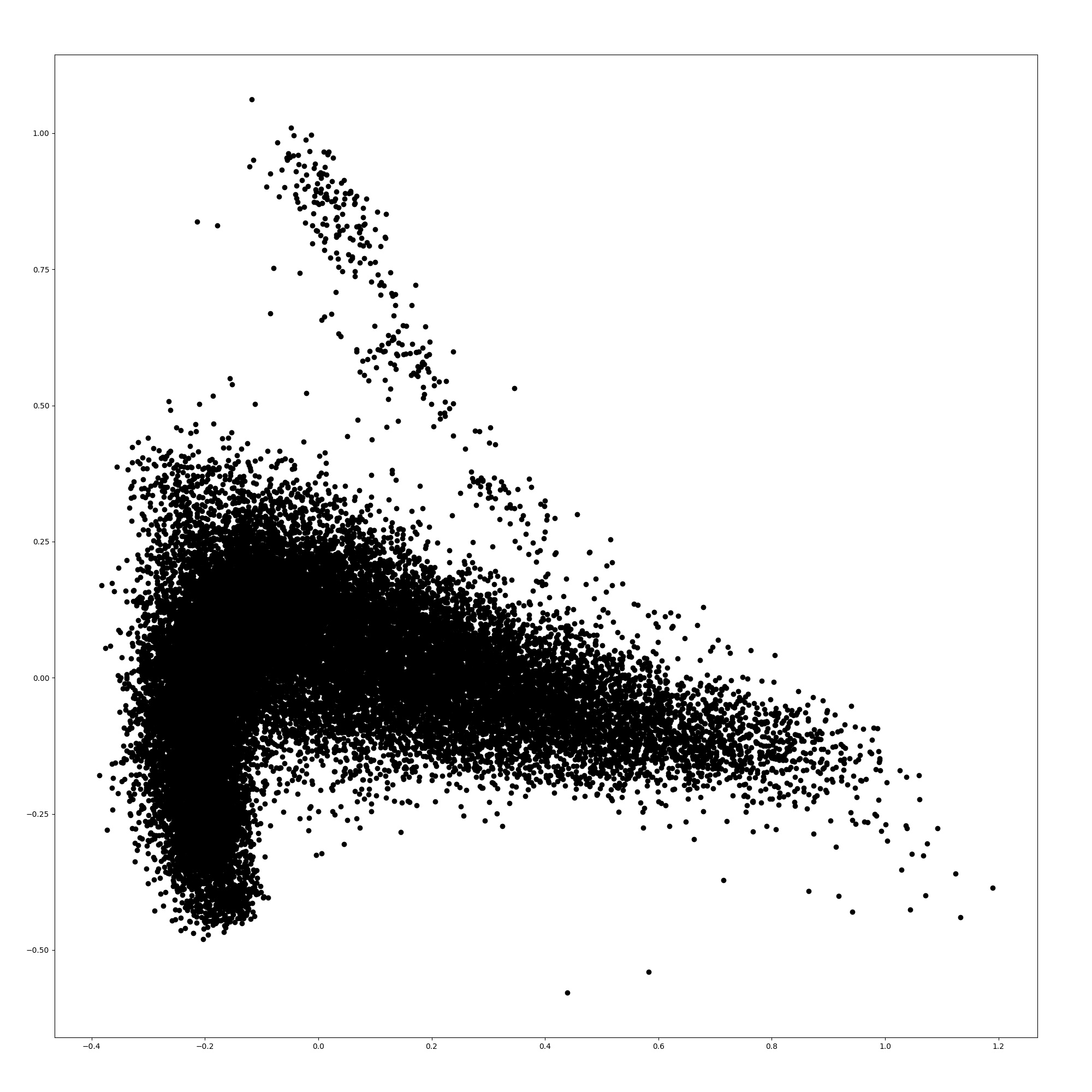
Here, we show the word embeddings of the models with principal component analysis. Figures 3, 3, 5, and 5 are scatter plots of the word embeddings of BERT-base-cased, the model additionally pre-trained with TAPT, the model additionally pre-trained with TAPER, and BioBERT.
The figures show that the word embeddings of BERT-base-cased and TAPT resemble each other. The average distance between the embeddings of BERT-base-cased and TAPT among all words is 0.0576, although the distance between the embeddings of BERT-base-cased and TAPTER is 0.172. Therefore, the additional pre-training of language modeling alone does not adapt the static word embeddings to the biomedical domain unlike TAPTER. TAPTER and BioBERT have dissimilar word embedding distributions to that of BERT-base-cased.