:
\theoremsep
\jmlrvolume
\firstpageno1
\jmlryear2022
\jmlrworkshopMachine Learning for Health (ML4H) 2022
Turning Silver into Gold: Domain Adaptation with Noisy Labels for Wearable Cardio-Respiratory Fitness Prediction
Abstract
Deep learning models have shown great promise in various healthcare applications. However, most models are developed and validated on small-scale datasets, as collecting high-quality (gold-standard) labels for health applications is often costly and time-consuming. As a result, these models may suffer from overfitting and not generalize well to unseen data. At the same time, an extensive amount of data with imprecise labels (silver-standard) is starting to be generally available, as collected from inexpensive wearables like accelerometers and electrocardiography sensors. These currently underutilized datasets and labels can be leveraged to produce more accurate clinical models. In this work, we propose UDAMA, a novel model with two key components: Unsupervised Domain Adaptation and Multi-discriminator Adversarial training, which leverage noisy data from source domain (the silver-standard dataset) to improve gold-standard modeling. We validate our framework on the challenging task of predicting lab-measured maximal oxygen consumption (VO2max), the benchmark metric of cardio-respiratory fitness, using free-living wearable sensor data from two cohort studies as inputs. Our experiments show that the proposed framework achieves the best performance of corr = 0.665 0.04, paving the way for accurate fitness estimation at scale.
keywords:
Domain Adaptation, Deep Learning, Distribution Shift, Cardio-Respiratory Fitness1 Introduction
Deep learning (DL) has been widely applied to many healthcare applications, such as sleep stage classification, stress detection, and fitness prediction (Yildirim et al., 2019; Jaques et al., 2017; Sakr et al., 2018). Collecting high-quality labels (i.e., gold-standard) for healthcare applications may require extensive efforts and can be particularly time-consuming: as a consequence, most of the existing datasets are small-scale. Developing DL models with such small-scale datasets often leads to poor performance and lack of model generalization (Raschka, 2018). At the same time, large-scale datasets with imprecise labels (i.e., silver-standard datasets) are generally available from inexpensive wearable sensors. For instance, polysomnography (PSG) sensors are used as the gold-standard measurement for sleep stages monitoring. PSG is mainly utilized in a controlled environment since they it requires dedicated equipment and the involvement of clinicians. As a result, such high-quality (gold-standard) datasets are usually small-scale (O’Reilly et al., 2014). Recently, Electrocardiogram (ECG) sensors embedded in wearable devices have gained momentum due to their affordability and high portability, leading to the generation of less-accurate but large-scale (sliver-standard) datasets (Lee et al., 2022).
High quality labels are crucial for the development and validation of robust clinical models. The labels of silver-standard datasets often contain estimation noise due to less accurate collection scheme and, as they are collected from larger populations, they are characterized by distribution shifts, which makes validation against gold-standard data difficult. Therefore, a natural question arises: Can we leverage noisy large-scale sliver-standard datasets to improve DL model validation on gold-standard datasets?
Transfer learning (TF) is the natural candidate for this problem (Xu et al., 2020). However, it is difficult for it to handle the problem where the distribution between two tasks is not similar. Recently, domain adaptation (DA) has achieved some initial success on solving the problem of the distribution mismatch between the source and target domains (Patricia and Caputo, 2014). Specifically, adversarial-based DAs demonstrated state-of-the-art performance on reducing the difference between data distributions (Tzeng et al., 2017; Hassan Pour Zonoozi and Seydi, 2022), for example for medical imaging tasks (Venkataramani et al., 2018). However, adversarial-based DAs mainly focus on discriminating the source and target domains as a binary classification task but ignore the fine-grained information that resides within the distribution shifts. To this end, in this paper we introduce a multi-discriminator model to simultaneously learn the coarse-grained and fine-grained domain information under the DA scheme.
In particular, we focus on the challenging health application of cardiorespiratory fitness (CRF) prediction employing maximal oxygen consumption (VO2max) as the ground truth. CRF is a vital indicator of Cardiovascular disease (CVD) (Laukkanen et al., 2001), a leading cause of death globally (Kaptoge et al., 2019). Collecting gold-standard VO2max labels through maximal exercise tests is difficult because it requires participants to reach exhaustion while on a treadmill, using a face mask with a computerized gas analysis system to monitor ventilation and expired gas fractions, as shown in Figure 1. This is not practical for particular groups, such as the elderly, for instance. Therefore, an alternative submaximal VO2max test (Gonzales et al., 2020) promises to capture fitness levels with lower accuracy. Although the gold-standard and silver-standard data capture similar information, they suffer from distribution shift.
In our work, we propose a novel unsupervised domain adaptation framework to improve the gold-standard fitness prediction by leveraging large-scale noisy VO2max data.
2 Methods
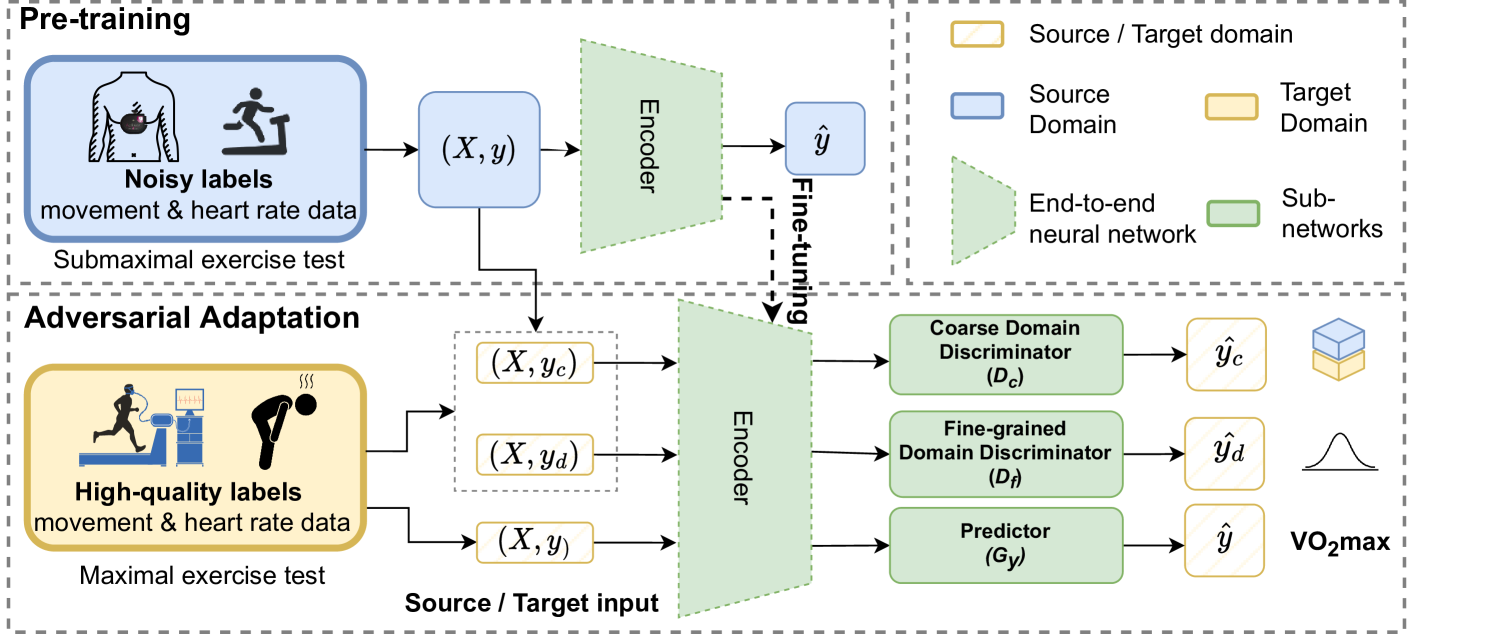
This work introduces a novel unsupervised domain adaptation framework that utilizes multi-discriminator during adversarial training. Herein, the section discussed details of our framework.
The overall model architecture and multi-discriminator training scheme are shown in Figure 1. In general, our framework consists of two training stages, pre-training on the source domain and an adversarial-based training stage on the (target domain). In the first stage, we build a pre-trained model with noisy health-related labels in a fully supervised setting. Specifically, the neural network is built through multiple stacked bidirectional Gated Recurrent Unit network (GRU) layers to extract time-series features’ embedding. After that, Multilayer Perceptron (MLP) layers are followed to extract associated metadata representation.
After pre-training, we fine-tune the large-scale feature embedding on after incorporating samples from . Besides, a novel multiple discriminator-based adversarial domain adaptation learning scheme is trained to distinguish domain labels and and estimate .
Multi-discriminator Adversarial Training. Our framework uses a combination of coarse-grained and fine-grained discriminators to aid the adversarial training for regression tasks. The coarse-grained discriminator () is similar to other regression DA (de Mathelin et al., 2020; Zhao et al., 2018) and follows the DANN style. In particular, mainly focus on discriminating rough domains, which uses a binary domain classifier to discriminate the source of data points (where 0 represents the data comes from the and 1 from ). However, the simple binary classes cannot fully represent the data label distribution of each domain. Therefore, we add a more fine-grained discriminator () to discriminate general distribution differences to aid the adversarial training.
For adversarial training, the shared encoder first leverages the pre-trained model and extract general feature. Then discriminators are trained to differentiate the domain labels. For , we optimize , a standard cross-entropy loss, to classify the coarse domain source labels . We can force the extractor to learn a general feature by maximizing such divergence. After that, a further maximizes the nuance changes among the sample and updates the discriminator by discriminating the domain distribution label using Gaussian Negative Log-Likelihood (GLL) loss (Paszke et al., 2019). After that, this multiple two-game adversarial training between the feature encoder and discriminator finally converges into balance. As a result, the feature extractor learns cross-domain knowledge from the multiple discriminators and thus benefits from the main regression prediction task. The whole framework is optimized by the total loss , which is defined as:
| (1) |
We use to minimize the loss of the predictor while maximizing the loss of domain discriminators. In detail, is used to scale down the predictor loss to the same level as predictors, and control the relative weight of the discriminator loss, and + = 1.
3 Dataset
Datasets. The source domain comes from the silver-standard measurement study (Fenland), including 12,425 participants. It contains a combined heart rate and movement signal from chest ECG sensor Actiheart and noisy VO2max labels collected from submaximal exercise tests (Lindsay et al., 2019). The target domain represents the gold-standard measurement dataset BBVS, which is a subset of 191 participants from the Fenland study with directly measured ground-truth VO2max (Gonzales et al., 2021) during the maximal exercise tests. In the BBVS study, participants need to wear a face mask to measure respiratory gas measurements (Rietjens et al., 2001) to experience exhaustion tests. Further, movement and heart rate are collected using the Actiheart (CamNtech, Papworth, UK) sensor. The details of features and the dataset are in Appendix A
Training Strategy. We evaluate UDAMA on these two datasets by first pre-training a model on the Fenland. Second, we develop the adversarial training framework with multi-discriminators on the BVS . For each domain, feature choices are same as previous study (Spathis et al., 2022). After the adversarial adaptation, we predict VO2max on the held-out test set of BVS using 3-fold cross-validation using UDAMA. Within each fold, the dataset is split into 70% training and 30% testing consisting only of target domain samples.
4 Results and Discussion
| Method | R2 | Corr | MSE | MAE |
| Train from scratch | -0.096 0.100 | 0.007 0.250 | 58.048 10.061 | 6.336 0.621 |
| Transfer learning (TF) | 0.283 0.037 | 0.621 0.012 | 35.399 5.91 | 4.744 0.433 |
| UDAMA (ours) | 0.392 0.07 | 0.665 0.04 | 30.794 6.805 | 4.442 0.328 |
4.1 VO2max prediction
The comparison of the proposed domain adaptation framework and baseline approach for VO2max prediction is shown in Table 1. The proposed framework outperforms all baselines for this prediction task and yields the best results among all evaluation metrics. We observe that the correlation (Corr) outperforms the basic transfer learning methods by 7.1%, the MSE increases by 13.0%, and R2 improves by 38.5% compared to the baseline Transfer Learning method.
4.2 Domain shift
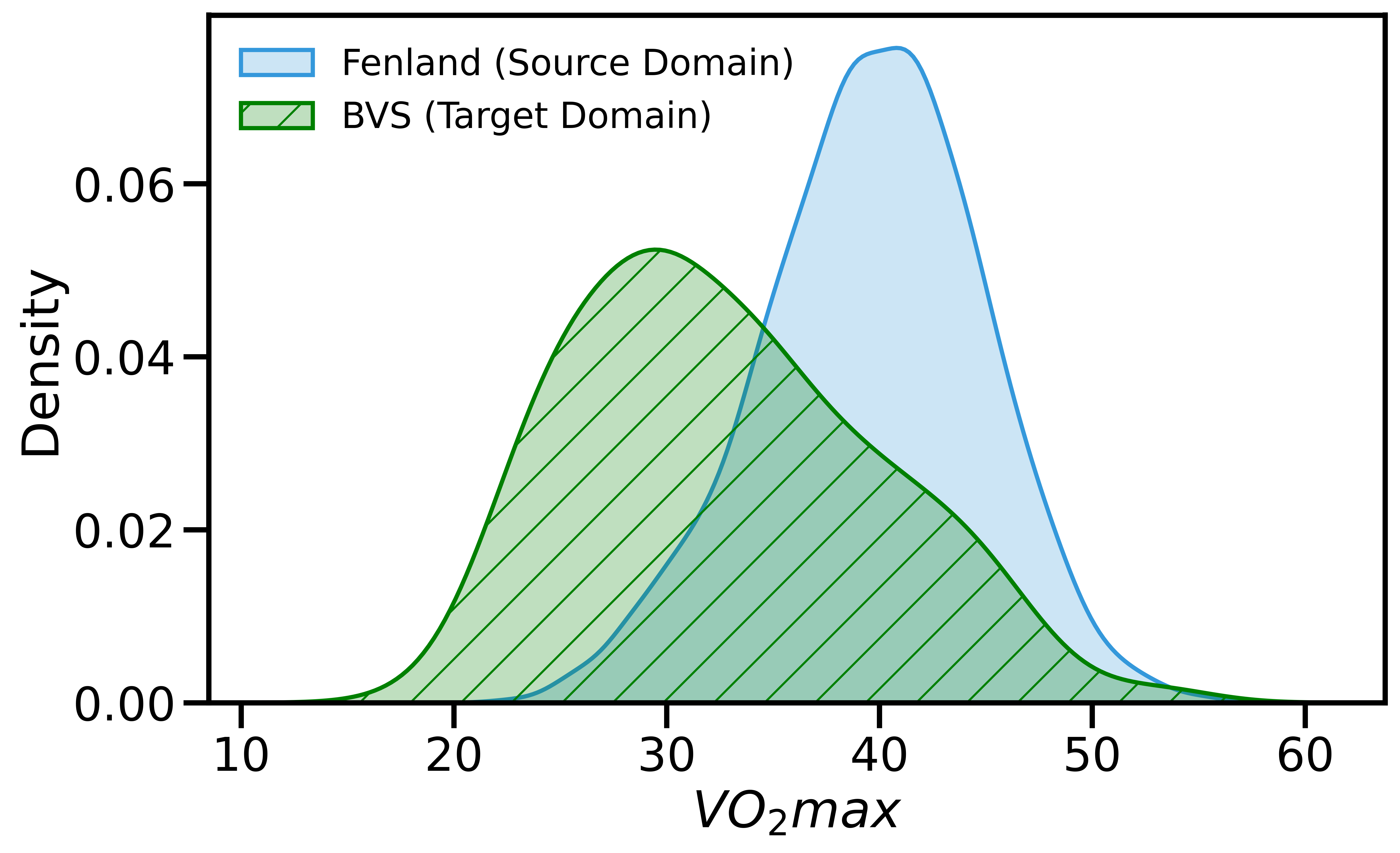
We examined whether UDAMA can effectively solve the domain shift problem. We observe that our proposed domain adaptation framework can effectively alleviate the problem. Figure 2 shows that the dataset (i.e., Fenland) shares a different VO2max underlying distribution compared to BBVS. Our results in Figure 3 show that our framework learns the small dataset distribution during the adaptation phase and achieves promising results compared with other methods. To examine the distance between domains, we use the Hellinger Distance (HD) to calculate the similarity of distributions between the prediction and the ground truth. Specifically, our framework’s prediction of fitness level lies in the same range as the ground truth, where the mean of the UDAMA is 33.96, and the mean of BBVS is 32.95. Besides, distribution ties close compared to baseline methods, where the normalized HD for UDAMA is 0.188, and HD for TF is 0.264. These results indicate that our framework can effectively alleviate the distribution shift problem of the VO2max prediction task. As a result, we reduce the difference between noisy silver-standard data and gold-standard data and improve the gold-standard model performance.
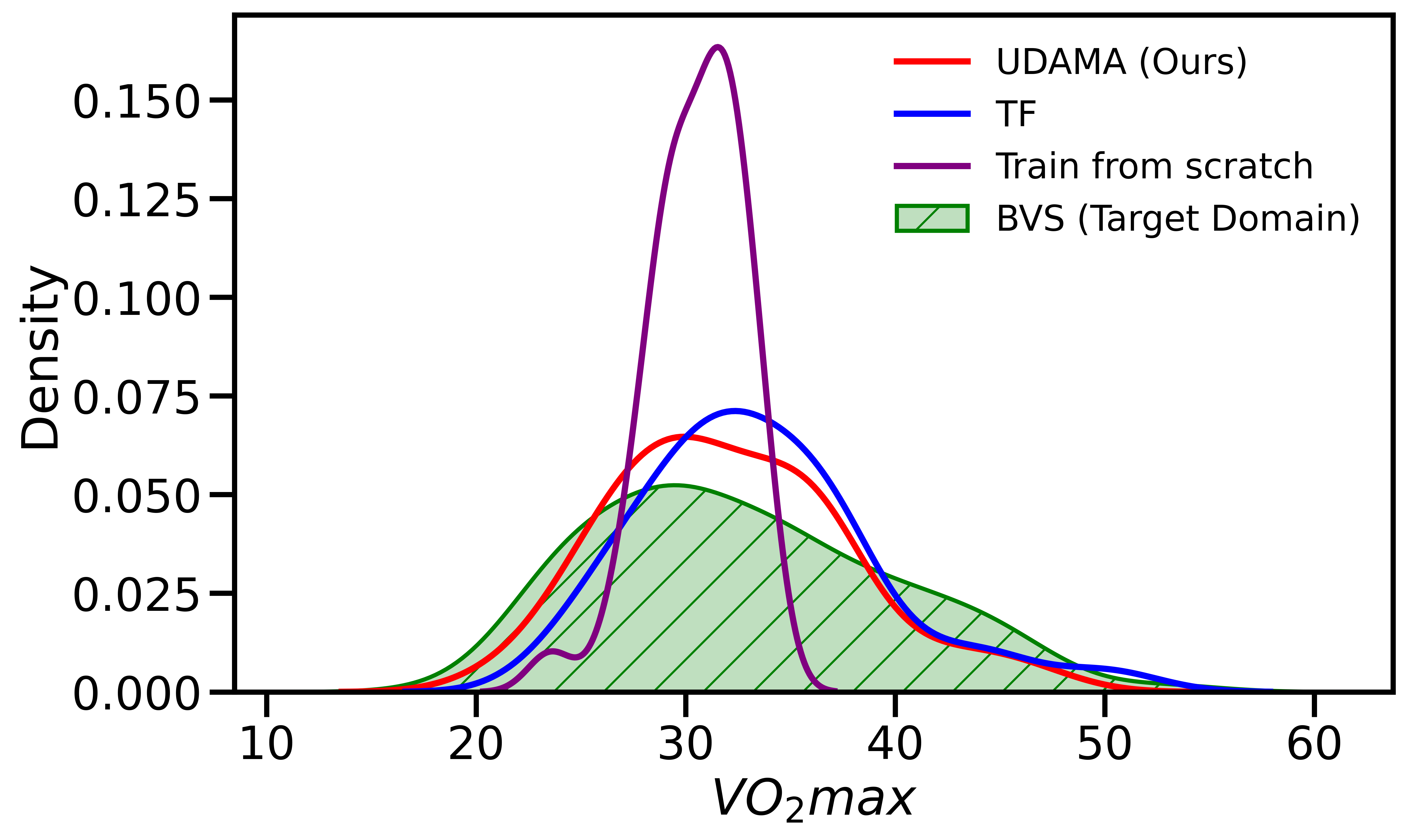
5 Conclusion
In this work, we proposed the unlabeled domain adaptation via multi-discriminator adversarial training framework (UDAMA) to address the problem of training datasets with various levels of label noise and distribution shifts. By leveraging a large-scale noisy silver-standard dataset and adversarial-based domain adaptation, our proposed method alleviates the domain shit problem and improves the performance on the challenging VO2max prediction task. Future work includes generalizing UDAMA to other healthcare applications, especially regression tasks involving high-dimensional time-series or wearable-sourced data.
This work is supported by ERC through Project 833296 (EAR) and by Nokia Bell Labs through a donation.
References
- de Mathelin et al. (2020) Antoine de Mathelin, Guillaume Richard, Francois Deheeger, Mathilde Mougeot, and Nicolas Vayatis. Adversarial weighting for domain adaptation in regression, 2020. URL https://arxiv.org/abs/2006.08251.
- Gonzales et al. (2020) Tomas Gonzales, Kate Westgate, Stefanie Hollidge, Tim Lindsay, Justin Jeon, and Soren Brage. Estimating maximal oxygen consumption from heart rate response to submaximal ramped treadmill test, 02 2020.
- Gonzales et al. (2021) Tomas I. Gonzales, Kate Westgate, Tessa Strain, Stefanie Hollidge, Justin Jeon, Dirk L. Christensen, Jorgen Jensen, Nicholas J. Wareham, and Søren Brage. Cardiorespiratory fitness assessment using risk-stratified exercise testing and dose-response relationships with disease outcomes. Scientific Reports, 11(1):15315, Jul 2021. ISSN 2045-2322. 10.1038/s41598-021-94768-3. URL https://doi.org/10.1038/s41598-021-94768-3.
- Hassan Pour Zonoozi and Seydi (2022) Mahta Hassan Pour Zonoozi and Vahid Seydi. A survey on adversarial domain adaptation. Neural Processing Letters, 08 2022. 10.1007/s11063-022-10977-5.
- Jaques et al. (2017) Natasha Jaques, Ognjen (Oggi) Rudovic, Sara Taylor, Akane Sano, and Rosalind Picard. Predicting tomorrow’s mood, health, and stress level using personalized multitask learning and domain adaptation. In Neil Lawrence and Mark Reid, editors, Proceedings of IJCAI 2017 Workshop on Artificial Intelligence in Affective Computing, volume 66 of Proceedings of Machine Learning Research, pages 17–33. PMLR, 20 Aug 2017. URL https://proceedings.mlr.press/v66/jaques17a.html.
- Kaptoge et al. (2019) Stephen Kaptoge, Lisa Pennells, Dirk De Bacquer, Marie Therese Cooney, Maryam Kavousi, Gretchen Stevens, Leanne Margaret Riley, Stefan Savin, Taskeen Khan, Servet Altay, Philippe Amouyel, Gerd Assmann, Steven Bell, Yoav Ben-Shlomo, Lisa Berkman, Joline W. Beulens, Cecilia Björkelund, Michael Blaha, Dan G. Blazer, Thomas Bolton, Ruth Bonita Beaglehole, Hermann Brenner, Eric J. Brunner, Edoardo Casiglia, Parinya Chamnan, Yeun-Hyang Choi, Rajiv Chowdry, Sean Coady, Carlos J. Crespo, Mary Cushman, Gilles R. Dagenais, Ralph B. D’Agostino Sr, Makoto Daimon, Karina W. Davidson, Gunnar Engström, Ian Ford, John Gallacher, Ron T. Gansevoort, Thomas Andrew Gaziano, Simona Giampaoli, Greg Grandits, Sameline Grimsgaard, Diederick E. Grobbee, Vilmundur Gudnason, Qi Guo, Hanna Tolonen, Steve Humphries, Hiroyasu Iso, J. Wouter Jukema, Jussi Kauhanen, Andre Pascal Kengne, Davood Khalili, Wolfgang Koenig, Daan Kromhout, Harlan Krumholz, T. H. Lam, Gail Laughlin, Alejandro Marín Ibañez, Tom W. Meade, Karel G. M. Moons, Paul J. Nietert, Toshiharu Ninomiya, Børge G. Nordestgaard, Christopher O’Donnell, Luigi Palmieri, Anushka Patel, Pablo Perel, Jackie F. Price, Rui Providencia, Paul M. Ridker, Beatriz Rodriguez, Annika Rosengren, Ronan Roussel, Masaru Sakurai, Veikko Salomaa, Shinichi Sato, Ben Schöttker, Nawar Shara, Jonathan E. Shaw, Hee-Choon Shin, Leon A. Simons, Eleni Sofianopoulou, Johan Sundström, Henry Völzke, Robert B. Wallace, Nicholas J. Wareham, Peter Willeit, David Wood, Angela Wood, Dong Zhao, Mark Woodward, Goodarz Danaei, Gregory Roth, Shanthi Mendis, Oyere Onuma, Cherian Varghese, Majid Ezzati, Ian Graham, Rod Jackson, John Danesh, and Emanuele Di Angelantonio. World health organization cardiovascular disease risk charts: revised models to estimate risk in 21 global regions. The Lancet Global Health, 7(10):e1332–e1345, Oct 2019. ISSN 2214-109X. 10.1016/S2214-109X(19)30318-3. URL https://doi.org/10.1016/S2214-109X(19)30318-3.
- Laukkanen et al. (2001) Jari A. Laukkanen, Timo A. Lakka, Rainer Rauramaa, Raimo Kuhanen, Juha M. Venäläinen, Riitta Salonen, and Jukka T. Salonen. Cardiovascular Fitness as a Predictor of Mortality in Men. Archives of Internal Medicine, 161(6):825–831, 03 2001. URL https://doi.org/10.1001/archinte.161.6.825.
- Lee et al. (2022) Harlin Lee, Boyue Li, Shelly DeForte, Mark Splaingard, Yungui Huang, Yuejie Chi, and Simon Linwood. A large collection of real-world pediatric sleep studies. Scientific Data, 9, 07 2022. 10.1038/s41597-022-01545-6.
- Lindsay et al. (2019) Tim Lindsay, Kate Westgate, Katrien Wijndaele, Stefanie Hollidge, Nicola Kerrison, Nita Forouhi, Simon Griffin, Nick Wareham, and Soren Brage. Descriptive epidemiology of physical activity energy expenditure in uk adults (the fenland study). International Journal of Behavioral Nutrition and Physical Activity, 16, 12 2019. 10.1186/s12966-019-0882-6.
- O’Reilly et al. (2014) Christian O’Reilly, Nadia Gosselin, and Julie Carrier. Montreal archive of sleep studies: an open-access resource for instrument benchmarking and exploratory research. Journal of sleep research, 23, 06 2014. 10.1111/jsr.12169.
- Paszke et al. (2019) Adam Paszke, Sam Gross, Francisco Massa, Adam Lerer, James Bradbury, Gregory Chanan, Trevor Killeen, Zeming Lin, Natalia Gimelshein, Luca Antiga, Alban Desmaison, Andreas Köpf, Edward Yang, Zach DeVito, Martin Raison, Alykhan Tejani, Sasank Chilamkurthy, Benoit Steiner, Lu Fang, Junjie Bai, and Soumith Chintala. Pytorch: An imperative style, high-performance deep learning library, 2019. URL https://arxiv.org/abs/1912.01703.
- Patricia and Caputo (2014) Novi Patricia and Barbara Caputo. Learning to learn, from transfer learning to domain adaptation: A unifying perspective. In 2014 IEEE Conference on Computer Vision and Pattern Recognition, pages 1442–1449, 2014. 10.1109/CVPR.2014.187.
- Raschka (2018) Sebastian Raschka. Model evaluation, model selection, and algorithm selection in machine learning. CoRR, abs/1811.12808, 2018. URL http://arxiv.org/abs/1811.12808.
- Rietjens et al. (2001) G. Rietjens, H Kuipers, A. Kester, and HA Keizer. Validation of a computerized metabolic measurement system (oxycon-pro®) during low and high intensity exercise1. International journal of sports medicine, 22:291–4, 06 2001. 10.1055/s-2001-14342.
- Sakr et al. (2018) Sherif Sakr, Radwa El Shawi, Amjad Ahmed, Waqas Qureshi, Clinton Brawner, Steven Keteyian, Michael Blaha, and Mouaz Al-Mallah. Using machine learning on cardiorespiratory fitness data for predicting hypertension: The henry ford exercise testing (fit) project. PloS one, 13:e0195344, 04 2018. 10.1371/journal.pone.0195344.
- Spathis et al. (2021) Dimitris Spathis, Ignacio Perez-Pozuelo, Soren Brage, Nicholas J. Wareham, and Cecilia Mascolo. Self-supervised transfer learning of physiological representations from free-living wearable data. In Proceedings of the Conference on Health, Inference, and Learning. ACM, apr 2021. 10.1145/3450439.3451863. URL https://doi.org/10.1145%2F3450439.3451863.
- Spathis et al. (2022) Dimitris Spathis, Ignacio Perez-Pozuelo, Tomas I. Gonzales, Soren Brage, Nicholas Wareham, and Cecilia Mascolo. Longitudinal cardio-respiratory fitness prediction through free-living wearable sensors, 2022. URL https://arxiv.org/abs/2205.03116.
- Stegle et al. (2008) Oliver Stegle, Sebastian V. Fallert, David J. C. MacKay, and Soren Brage. Gaussian process robust regression for noisy heart rate data. IEEE Transactions on Biomedical Engineering, 55(9):2143–2151, 2008. 10.1109/TBME.2008.923118.
- Tzeng et al. (2017) Eric Tzeng, Judy Hoffman, Kate Saenko, and Trevor Darrell. Adversarial discriminative domain adaptation, 2017. URL https://arxiv.org/abs/1702.05464.
- Venkataramani et al. (2018) Rahul Venkataramani, Hariharan Ravishankar, and Saihareesh Anamandra. Towards continuous domain adaptation for healthcare, 2018. URL https://arxiv.org/abs/1812.01281.
- Xu et al. (2020) Wen Xu, Jing He, and Yanfeng Shu. Transfer learning and deep domain adaptation. In Marco Antonio Aceves-Fernandez, editor, Advances and Applications in Deep Learning, chapter 3. IntechOpen, Rijeka, 2020. 10.5772/intechopen.94072. URL https://doi.org/10.5772/intechopen.94072.
- Yildirim et al. (2019) Özal Yildirim, Ulas Baloglu, and U Rajendra Acharya. A deep learning model for automated sleep stages classification using psg signals. International journal of environmental research and public health, 16:599, 02 2019. 10.3390/ijerph16040599.
- Zhao et al. (2018) Han Zhao, Shanghang Zhang, Guanhang Wu, José M. F. Moura, Joao P Costeira, and Geoffrey J Gordon. Adversarial multiple source domain adaptation. In S. Bengio, H. Wallach, H. Larochelle, K. Grauman, N. Cesa-Bianchi, and R. Garnett, editors, Advances in Neural Information Processing Systems, volume 31. Curran Associates, Inc., 2018. URL https://proceedings.neurips.cc/paper/2018/file/717d8b3d60d9eea997b35b02b6a4e867-Paper.pdf.
Appendix A Appendix
A.1 Problem Formulation and Notation
Here, we denote as the source domain containing silver-standard data and as the target domain, as shown in Figure 1. For each domain we assume samples, an input time-series sequence , the corresponding label is , where represents the coarse-grained binary domain label, denotes the fine-grained domain distribution label, and denotes the VO2max prediction. During the adaptation phase, we use multiple discriminators to distinguish . In particular, the coarse-grained domain discriminator is and the fine-grained domain discriminator is . Also, for the training process, we denote the feature encoder with , and the regression predictor with .
A.2 Dataset
All heart rate data collected during free-living conditions underwent pre-processing for noise filtering (Stegle et al., 2008). If participants had fewer than 72 hours of concurrent wear data (three full days of recording) or inadequate individual calibration data, they were eliminated from the study (treadmill test-based data). Non-wear periods were excluded from the analyses through non-wear detection procedures. This pre-processing algorithm discovered lengthy durations of non-physiological heart rate and extended periods of no movement reported by the device’s accelerometer (>90 minutes). We used the calculation 1 MET = 71 J/min/kg (3.5 ml O2 min1 kg1) to convert movement intensities into standard metabolic equivalent units (METs). These conversions were then used to classify intensity levels, with behaviors less than 1.5METs classed as sedentary, those between 3 and 6 METs as moderate to vigorous physical activity (MVPA), and those greater than 6METs as vigorous physical activity (VPA). Since time can greatly impact physical activities, we encoded the sensor timestamps using cyclical temporal features (Spathis et al., 2021).
Additionally, given the sensors’ high sampling rate (1 sample/minute) after matching the HR and Acceleration modalities, learning patterns from such a lengthy sequence (a week’s worth of sensor data contains more than 10,000 timesteps) is unfeasible, even with the most potent recurrent neural networks. Therefore, our models downsampled the sampling rate to 15 minutes and used the first 600 timesteps to represent each participant’s feature vector. Then each feature vector with 26 features combing time-series and metadata was put into various deep neural networks.